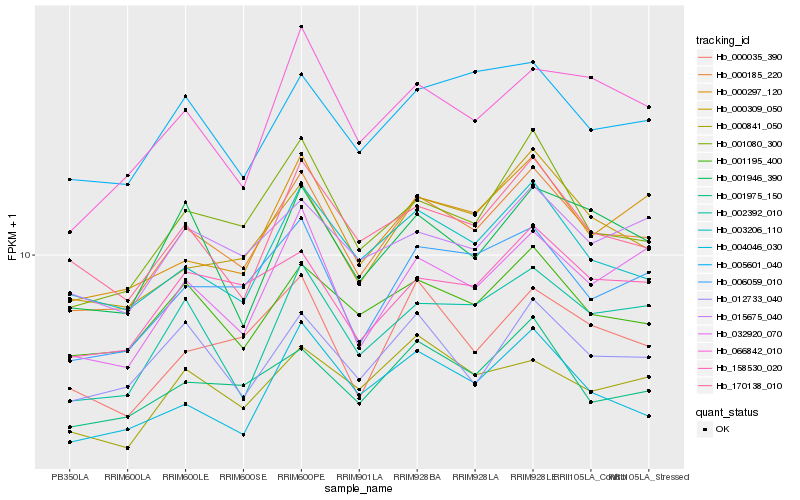
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000185_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 2 |
Hb_006059_010 |
0.0630738418 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 3 |
Hb_001195_400 |
0.0673160486 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 4 |
Hb_003206_110 |
0.0678151486 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 5 |
Hb_001080_300 |
0.068108351 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 6 |
Hb_000309_050 |
0.0685495784 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001975_150 |
0.0697724867 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 8 |
Hb_000297_120 |
0.0721518894 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 9 |
Hb_158530_020 |
0.0725578871 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 10 |
Hb_170138_010 |
0.0727418288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_032920_070 |
0.0762872695 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 12 |
Hb_015675_040 |
0.076862595 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 13 |
Hb_002392_010 |
0.0778180079 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 14 |
Hb_005601_040 |
0.0795715518 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 15 |
Hb_012733_040 |
0.0811483328 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 16 |
Hb_000035_390 |
0.0828637665 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 17 |
Hb_004046_030 |
0.0844008241 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 18 |
Hb_066842_010 |
0.0860335426 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000841_050 |
0.0860570623 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 20 |
Hb_001946_390 |
0.0864717936 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |