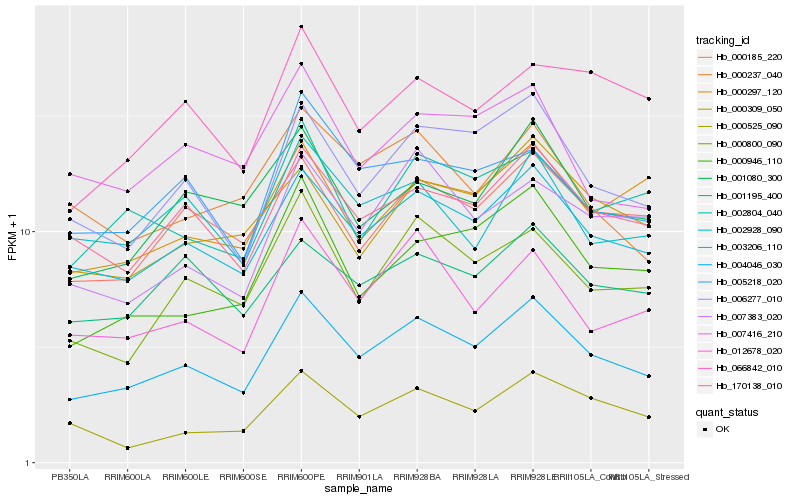
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004046_030 |
0.0 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 2 |
Hb_003206_110 |
0.0517741641 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 3 |
Hb_006277_010 |
0.0720132978 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 4 |
Hb_170138_010 |
0.0778877091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000185_220 |
0.0844008241 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 6 |
Hb_000800_090 |
0.0879134173 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 7 |
Hb_012678_020 |
0.0920581083 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001080_300 |
0.0940333014 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 9 |
Hb_000946_110 |
0.0945903817 |
- |
- |
GTP cyclohydrolase I, putative [Ricinus communis] |
| 10 |
Hb_005218_020 |
0.0967547572 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_002804_040 |
0.0972841082 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 12 |
Hb_000309_050 |
0.0974379699 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007383_020 |
0.0985237437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001195_400 |
0.1000633915 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 15 |
Hb_000297_120 |
0.1012937393 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 16 |
Hb_002928_090 |
0.1027716975 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 17 |
Hb_000525_090 |
0.1035508551 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 18 |
Hb_066842_010 |
0.1041523915 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000237_040 |
0.1062234403 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 20 |
Hb_007416_210 |
0.1062747425 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |