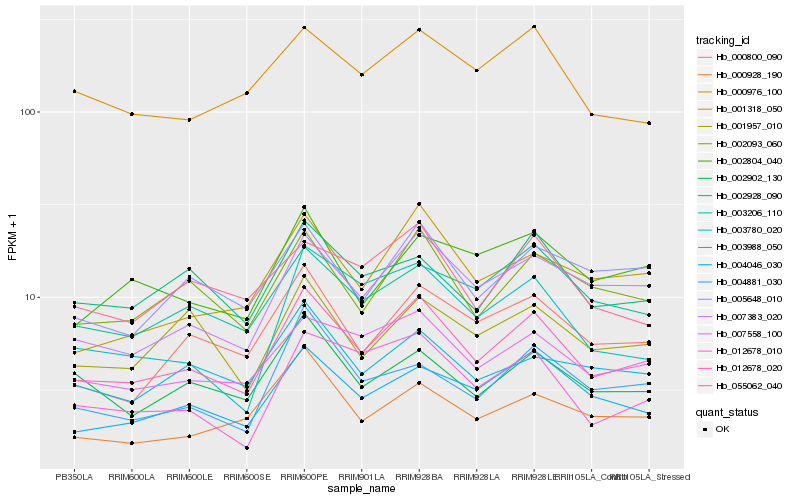
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012678_020 |
0.0 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007558_100 |
0.0855865174 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002093_060 |
0.0859458608 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_007383_020 |
0.0876389795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004046_030 |
0.0920581083 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 6 |
Hb_000800_090 |
0.0973474828 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003206_110 |
0.0994115479 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 8 |
Hb_001318_050 |
0.1007897334 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 9 |
Hb_005648_010 |
0.1020035273 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 10 |
Hb_003988_050 |
0.1028750736 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 11 |
Hb_002902_130 |
0.1030708063 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 12 |
Hb_012678_010 |
0.1034551169 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 13 |
Hb_002928_090 |
0.1046066828 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 14 |
Hb_000976_100 |
0.1055042221 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 15 |
Hb_000928_190 |
0.1066635228 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001957_010 |
0.1076727761 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 17 |
Hb_055062_040 |
0.1078658683 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 18 |
Hb_004881_030 |
0.1087954715 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002804_040 |
0.1097476445 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 20 |
Hb_003780_020 |
0.1103267034 |
- |
- |
PREDICTED: prolyl 4-hydroxylase subunit alpha-1 isoform X2 [Glycine max] |