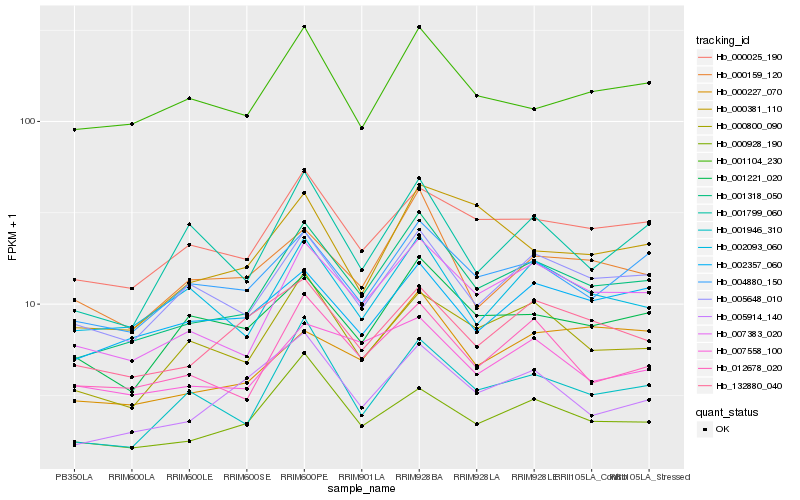
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_050 |
0.0 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 2 |
Hb_007383_020 |
0.0682184743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005648_010 |
0.0950378316 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 4 |
Hb_000025_190 |
0.0954150215 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 5 |
Hb_012678_020 |
0.1007897334 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004880_150 |
0.1008033279 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 7 |
Hb_000800_090 |
0.1010783789 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 8 |
Hb_005914_140 |
0.1030774434 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 9 |
Hb_001946_310 |
0.1035304424 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 10 |
Hb_001221_020 |
0.1049723749 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 11 |
Hb_000928_190 |
0.1082498211 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000227_070 |
0.1097431084 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 13 |
Hb_000159_120 |
0.1110282033 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 14 |
Hb_001104_230 |
0.1120908763 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 15 |
Hb_002093_060 |
0.1125978194 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_132880_040 |
0.1136663584 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002357_060 |
0.1142749339 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 18 |
Hb_000381_110 |
0.1167339589 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001799_060 |
0.1169036526 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 20 |
Hb_007558_100 |
0.1175378024 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |