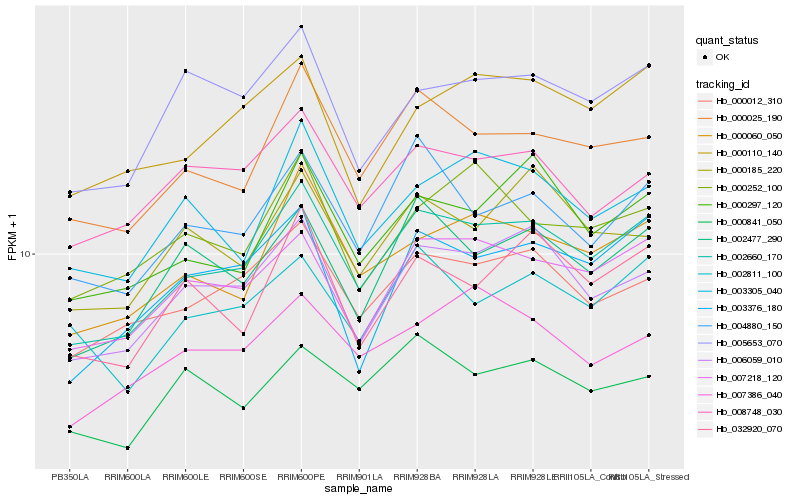
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002660_170 |
0.0 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_000025_190 |
0.0667451235 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_007218_120 |
0.0676461242 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 4 |
Hb_006059_010 |
0.069455339 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 5 |
Hb_000060_050 |
0.0705743526 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 6 |
Hb_000012_310 |
0.0722235729 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002477_290 |
0.078330372 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000297_120 |
0.0784134408 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 9 |
Hb_004880_150 |
0.0824534216 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 10 |
Hb_003376_180 |
0.0865195568 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 11 |
Hb_032920_070 |
0.0873131623 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 12 |
Hb_003305_040 |
0.0876066912 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 13 |
Hb_000110_140 |
0.0884199562 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 14 |
Hb_005653_070 |
0.0891091197 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 15 |
Hb_002811_100 |
0.0918117292 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 16 |
Hb_007386_040 |
0.0919697784 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 9, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_008748_030 |
0.0925746466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000252_100 |
0.0944771858 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 19 |
Hb_000185_220 |
0.095277526 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 20 |
Hb_000841_050 |
0.0955139521 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |