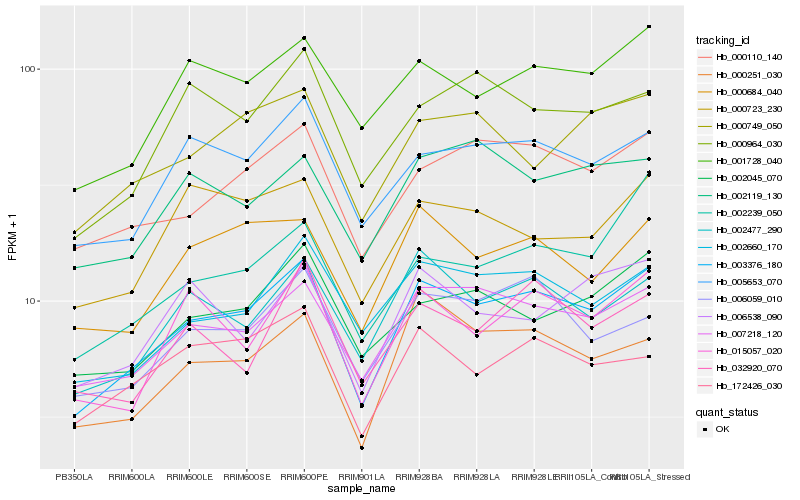
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_180 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 2 |
Hb_007218_120 |
0.0747669543 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000110_140 |
0.0842568983 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 4 |
Hb_002477_290 |
0.0851889341 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_005653_070 |
0.0854877515 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 6 |
Hb_002660_170 |
0.0865195568 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 7 |
Hb_001728_040 |
0.0870849293 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 8 |
Hb_000684_040 |
0.0917859466 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 9 |
Hb_000251_030 |
0.0919661567 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_015057_020 |
0.0926159134 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 11 |
Hb_002045_070 |
0.0950952395 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 12 |
Hb_006059_010 |
0.0963679853 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 13 |
Hb_000749_050 |
0.0987858139 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 14 |
Hb_172426_030 |
0.0989476591 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002239_050 |
0.1014451432 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000964_030 |
0.1023343807 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 17 |
Hb_000723_230 |
0.1024517876 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 18 |
Hb_006538_090 |
0.1044597565 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 19 |
Hb_032920_070 |
0.1047216579 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 20 |
Hb_002119_130 |
0.1047959456 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |