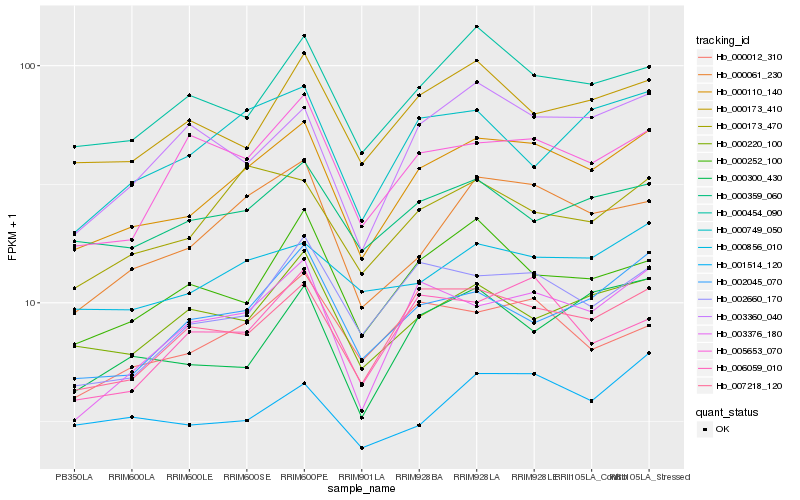
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_140 |
0.0 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 2 |
Hb_007218_120 |
0.0725906702 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000061_230 |
0.0733636237 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 4 |
Hb_003376_180 |
0.0842568983 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 5 |
Hb_000454_090 |
0.0858029641 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 6 |
Hb_000173_470 |
0.0864325401 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_002660_170 |
0.0884199562 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 8 |
Hb_002045_070 |
0.0897356361 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 9 |
Hb_000856_010 |
0.0898814812 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 10 |
Hb_000173_410 |
0.0923716347 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 11 |
Hb_000220_100 |
0.0925892909 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 [Jatropha curcas] |
| 12 |
Hb_000012_310 |
0.0931691645 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000359_060 |
0.0945796753 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 14 |
Hb_000749_050 |
0.0954859407 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 15 |
Hb_000252_100 |
0.0958121116 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 16 |
Hb_006059_010 |
0.0965342676 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 17 |
Hb_003360_040 |
0.0966231152 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 18 |
Hb_005653_070 |
0.0973888235 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 19 |
Hb_000300_430 |
0.0977349233 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 20 |
Hb_001514_120 |
0.0977610675 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |