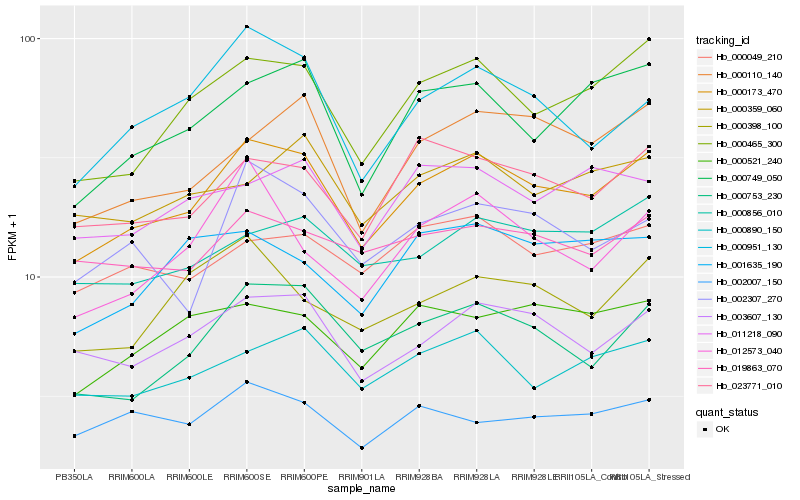
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_470 |
0.0 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_000465_300 |
0.0655991153 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 3 |
Hb_003607_130 |
0.074908273 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000753_230 |
0.0764286709 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000749_050 |
0.0823000315 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 6 |
Hb_023771_010 |
0.0826370516 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000890_150 |
0.0837865169 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Topors isoform X1 [Jatropha curcas] |
| 8 |
Hb_000110_140 |
0.0864325401 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 9 |
Hb_000856_010 |
0.0880307795 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 10 |
Hb_000951_130 |
0.0889743882 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 11 |
Hb_000521_240 |
0.0902045892 |
- |
- |
PREDICTED: uncharacterized protein LOC105650339 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000049_210 |
0.0911299455 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 13 |
Hb_019863_070 |
0.0941158047 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002307_270 |
0.0948911964 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 15 |
Hb_000359_060 |
0.0956270423 |
- |
- |
PREDICTED: WD repeat-containing protein 26-like [Jatropha curcas] |
| 16 |
Hb_000398_100 |
0.0965318017 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_011218_090 |
0.0972009781 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 18 |
Hb_012573_040 |
0.0978467118 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 19 |
Hb_002007_150 |
0.0984119223 |
- |
- |
- |
| 20 |
Hb_001635_190 |
0.0990622478 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |