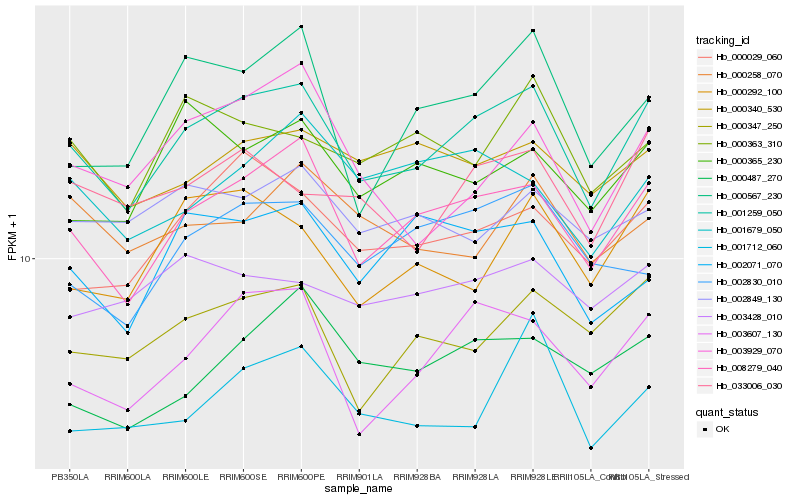
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001259_050 |
0.0 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 2 |
Hb_008279_040 |
0.0411421109 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_003607_130 |
0.0626428235 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000029_060 |
0.0691166706 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 5 |
Hb_033006_030 |
0.0744744076 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040-like [Jatropha curcas] |
| 6 |
Hb_000347_250 |
0.0757449631 |
- |
- |
PREDICTED: uncharacterized protein LOC105133971 [Populus euphratica] |
| 7 |
Hb_000567_230 |
0.0776368168 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 8 |
Hb_000487_270 |
0.0794665497 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 9 |
Hb_000340_530 |
0.0798415181 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 10 |
Hb_002830_010 |
0.079865368 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_001712_060 |
0.0801160034 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 12 |
Hb_000292_100 |
0.0801241527 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 13 |
Hb_000363_310 |
0.0802788973 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 14 |
Hb_003929_070 |
0.0811089936 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 15 |
Hb_003428_010 |
0.0814390039 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 16 |
Hb_002071_070 |
0.0816658604 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000365_230 |
0.0818532897 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 18 |
Hb_001679_050 |
0.082297476 |
- |
- |
PREDICTED: exocyst complex component EXO84B [Jatropha curcas] |
| 19 |
Hb_002849_130 |
0.082794889 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 20 |
Hb_000258_070 |
0.0832273247 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |