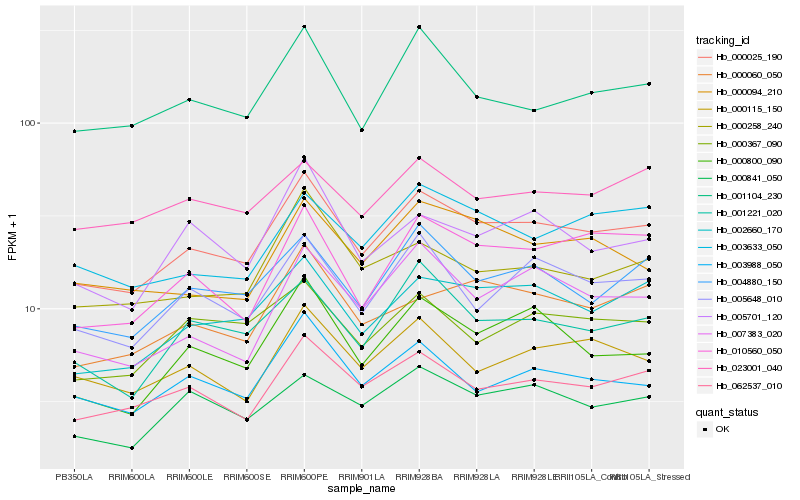
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_190 |
0.0 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_062537_010 |
0.0641081544 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 3 |
Hb_002660_170 |
0.0667451235 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_004880_150 |
0.073872905 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 5 |
Hb_005648_010 |
0.0774303 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 6 |
Hb_003988_050 |
0.0784563162 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 7 |
Hb_000115_150 |
0.0801991992 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 8 |
Hb_000060_050 |
0.0807902292 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 9 |
Hb_001104_230 |
0.0814955435 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 10 |
Hb_001221_020 |
0.0816353168 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 11 |
Hb_010560_050 |
0.0822295533 |
- |
- |
PREDICTED: acyl-protein thioesterase 1-like [Jatropha curcas] |
| 12 |
Hb_000800_090 |
0.0829618573 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000258_240 |
0.0837588815 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 14 |
Hb_000841_050 |
0.0839590338 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 15 |
Hb_000367_090 |
0.0855190369 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 16 |
Hb_007383_020 |
0.0865848753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003633_050 |
0.0879255697 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 18 |
Hb_000094_210 |
0.0884142989 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 19 |
Hb_005701_120 |
0.0898107972 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 20 |
Hb_023001_040 |
0.0908355217 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |