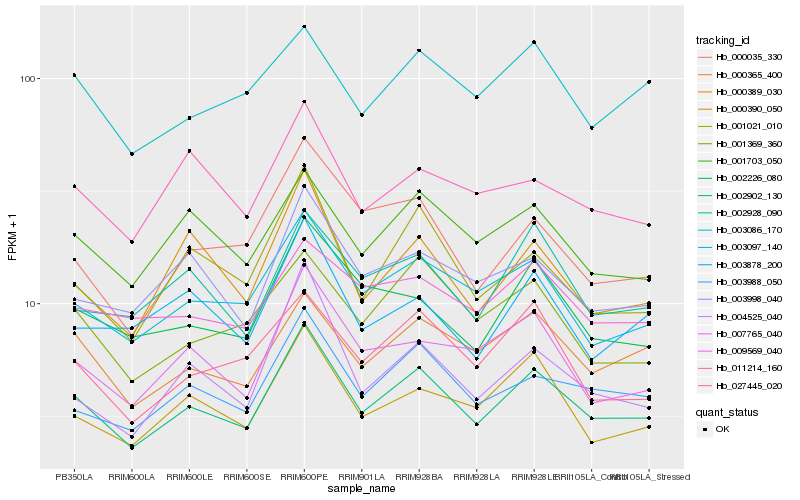
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002902_130 |
0.0 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 2 |
Hb_003988_050 |
0.0720652274 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 3 |
Hb_003097_140 |
0.0738726817 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 4 |
Hb_000365_400 |
0.0813178291 |
- |
- |
metal ion transporter, putative [Ricinus communis] |
| 5 |
Hb_000390_050 |
0.0817191636 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 6 |
Hb_001703_050 |
0.085176192 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_000389_030 |
0.086578787 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 8 |
Hb_007765_040 |
0.086910754 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 9 |
Hb_003998_040 |
0.0886786957 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 10 |
Hb_001021_010 |
0.0888777048 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 11 |
Hb_002226_080 |
0.0898039927 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002928_090 |
0.0912335953 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 13 |
Hb_027445_020 |
0.0932800313 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 14 |
Hb_001369_360 |
0.0941187258 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 15 |
Hb_004525_040 |
0.0946210136 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 16 |
Hb_003086_170 |
0.0956118931 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 17 |
Hb_009569_040 |
0.096024746 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 18 |
Hb_003878_200 |
0.0962664274 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |
| 19 |
Hb_011214_160 |
0.0963305889 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 20 |
Hb_000035_330 |
0.0970067628 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |