| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
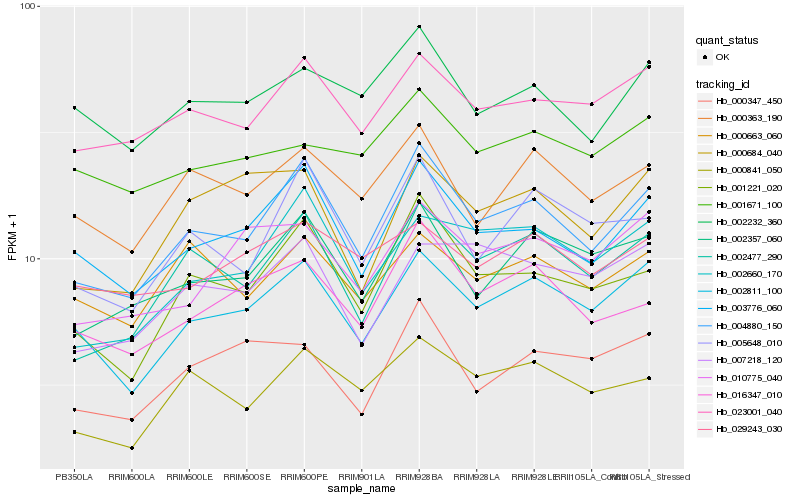
Hb_002811_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 2 |
Hb_004880_150 |
0.0704516919 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 3 |
Hb_003776_060 |
0.0708409697 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 4 |
Hb_023001_040 |
0.0821285403 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_010775_040 |
0.0851450328 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 6 |
Hb_000363_190 |
0.0859123707 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002357_060 |
0.089470315 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 8 |
Hb_002660_170 |
0.0918117292 |
- |
- |
PREDICTED: dystrophia myotonica WD repeat-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_000347_450 |
0.0934969257 |
- |
- |
PREDICTED: tRNA(His) guanylyltransferase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001221_020 |
0.0938832489 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 11 |
Hb_029243_030 |
0.0941231745 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002477_290 |
0.094383999 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000663_060 |
0.0946606263 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 14 |
Hb_002232_360 |
0.0949186348 |
- |
- |
PREDICTED: proline iminopeptidase isoform X2 [Jatropha curcas] |
| 15 |
Hb_007218_120 |
0.0954953308 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001671_100 |
0.0955029277 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 17 |
Hb_000684_040 |
0.0959918754 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 18 |
Hb_016347_010 |
0.097113359 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 19 |
Hb_005648_010 |
0.098442184 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 20 |
Hb_000841_050 |
0.0984839945 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |