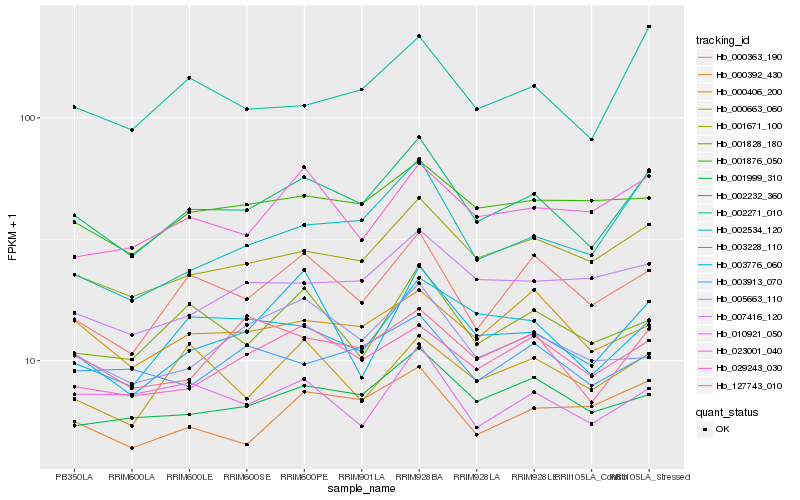
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_360 |
0.0 |
- |
- |
PREDICTED: proline iminopeptidase isoform X2 [Jatropha curcas] |
| 2 |
Hb_001671_100 |
0.0595598423 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 3 |
Hb_000363_190 |
0.0696969276 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001999_310 |
0.0710802731 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 5 |
Hb_029243_030 |
0.073758743 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000392_430 |
0.0764551376 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 7 |
Hb_001828_180 |
0.0774539073 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 8 |
Hb_001876_050 |
0.0781301432 |
- |
- |
hypothetical protein JCGZ_01286 [Jatropha curcas] |
| 9 |
Hb_127743_010 |
0.0790115181 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002271_010 |
0.0793315573 |
- |
- |
PREDICTED: 60S ribosomal protein L8 [Jatropha curcas] |
| 11 |
Hb_003228_110 |
0.0794074842 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 12 |
Hb_000406_200 |
0.0812469235 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_002534_120 |
0.0820307905 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |
| 14 |
Hb_007416_120 |
0.0831508274 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 15 |
Hb_005663_110 |
0.0832093399 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 16 |
Hb_023001_040 |
0.0839948167 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_003776_060 |
0.0853208957 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 18 |
Hb_000663_060 |
0.0864812948 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 19 |
Hb_003913_070 |
0.0879839028 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 20 |
Hb_010921_050 |
0.0881242372 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |