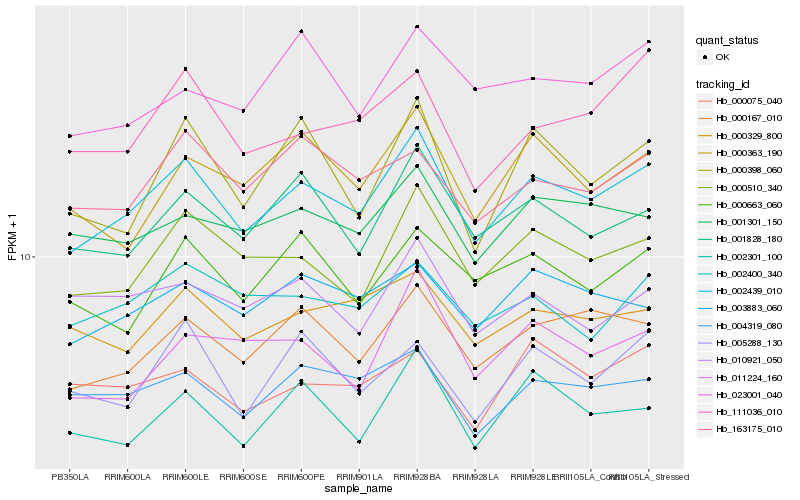
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002439_010 |
0.0 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 2 |
Hb_000510_340 |
0.0620986878 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 3 |
Hb_000167_010 |
0.0627371749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001828_180 |
0.0698664458 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 5 |
Hb_004319_080 |
0.0753958052 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 6 |
Hb_003883_060 |
0.0756924364 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001301_150 |
0.0760818345 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 8 |
Hb_000329_800 |
0.0773249859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000363_190 |
0.0799746497 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_163175_010 |
0.0801867285 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 11 |
Hb_000663_060 |
0.080899739 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 12 |
Hb_002400_340 |
0.0810841328 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 13 |
Hb_111036_010 |
0.0821750157 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 14 |
Hb_000398_060 |
0.0823105394 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 15 |
Hb_002301_100 |
0.0859547634 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 16 |
Hb_011224_160 |
0.086883714 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 17 |
Hb_010921_050 |
0.0876833482 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_023001_040 |
0.0879605589 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000075_040 |
0.0885324039 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 20 |
Hb_005288_130 |
0.0889714835 |
- |
- |
protein with unknown function [Ricinus communis] |