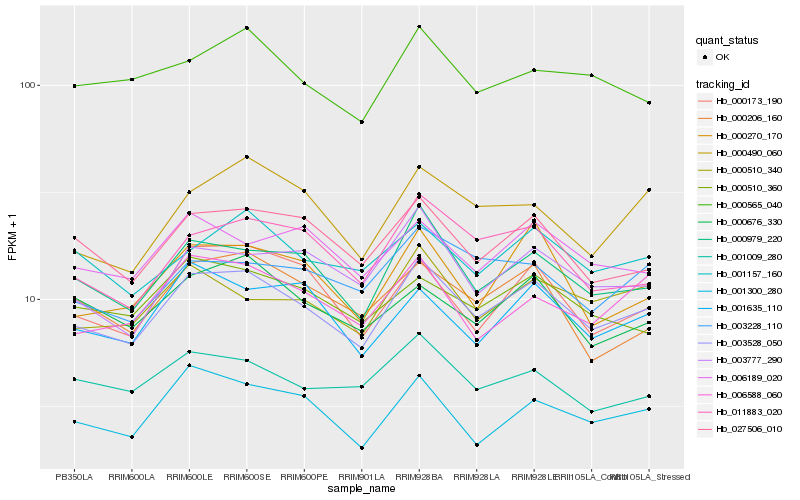
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003528_050 |
0.0 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 2 |
Hb_000979_220 |
0.0586760488 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 3 |
Hb_027506_010 |
0.0705016766 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 4 |
Hb_011883_020 |
0.0752730051 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 5 |
Hb_000510_340 |
0.0753300555 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 6 |
Hb_000173_190 |
0.0766975148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001300_280 |
0.0775575336 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 8 |
Hb_001157_160 |
0.0780494775 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003777_290 |
0.0784133518 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 10 |
Hb_001009_280 |
0.0789965964 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 11 |
Hb_006588_060 |
0.0792918703 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 12 |
Hb_000270_170 |
0.0804881978 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 13 |
Hb_000510_360 |
0.0812801493 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 14 |
Hb_006189_020 |
0.0822881323 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_000490_060 |
0.0826826626 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 16 |
Hb_001635_110 |
0.0827502887 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 17 |
Hb_000676_330 |
0.0828292699 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_003228_110 |
0.083251053 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 19 |
Hb_000206_160 |
0.0841803589 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000565_040 |
0.0841869407 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |