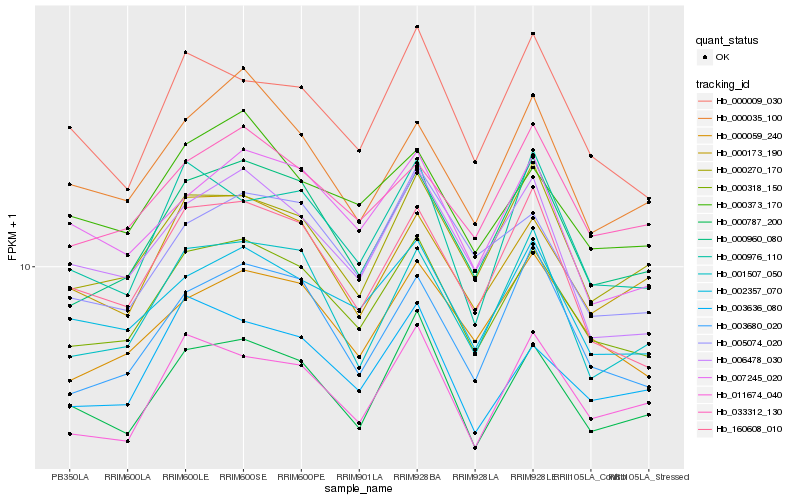
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_150 |
0.0 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 2 |
Hb_000960_080 |
0.0470032813 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002357_070 |
0.0569405759 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 4 |
Hb_000787_200 |
0.0626576494 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000059_240 |
0.0630233239 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 6 |
Hb_000270_170 |
0.0650770043 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 7 |
Hb_000173_190 |
0.0651882793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003680_020 |
0.0676818094 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_033312_130 |
0.0678674756 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 10 |
Hb_160608_010 |
0.0682808663 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 11 |
Hb_001507_050 |
0.0689160854 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_011674_040 |
0.0700080558 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003636_080 |
0.0700747377 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 14 |
Hb_000373_170 |
0.0705452446 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 15 |
Hb_006478_030 |
0.0706285676 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 16 |
Hb_007245_020 |
0.0733103043 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000976_110 |
0.0746444672 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 18 |
Hb_005074_020 |
0.0748733671 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000009_030 |
0.0753343371 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 20 |
Hb_000035_100 |
0.0759609801 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |