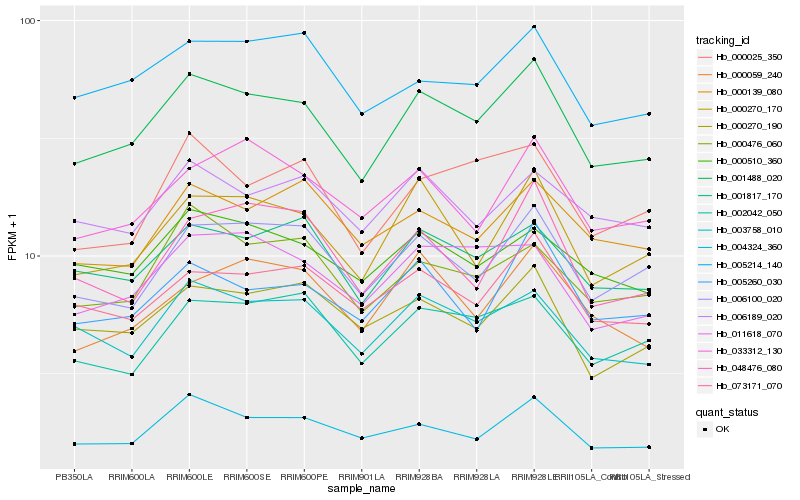
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_020 |
0.0 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 2 |
Hb_004324_360 |
0.0644706238 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000270_170 |
0.0669225118 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 4 |
Hb_001817_170 |
0.0671797637 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 5 |
Hb_006100_020 |
0.0676313399 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 6 |
Hb_003758_010 |
0.0680030148 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_073171_070 |
0.0698927061 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 8 |
Hb_011618_070 |
0.0751643368 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 9 |
Hb_002042_050 |
0.0758234646 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000270_190 |
0.0760878576 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 11 |
Hb_005214_140 |
0.0771152825 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 12 |
Hb_000510_360 |
0.078534331 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 13 |
Hb_000476_060 |
0.0786171672 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_005260_030 |
0.0786925972 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 15 |
Hb_000025_350 |
0.0804492744 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_033312_130 |
0.0812635521 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000139_080 |
0.0823694677 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006189_020 |
0.082483118 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 19 |
Hb_000059_240 |
0.0835008411 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 20 |
Hb_048476_080 |
0.0856778915 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |