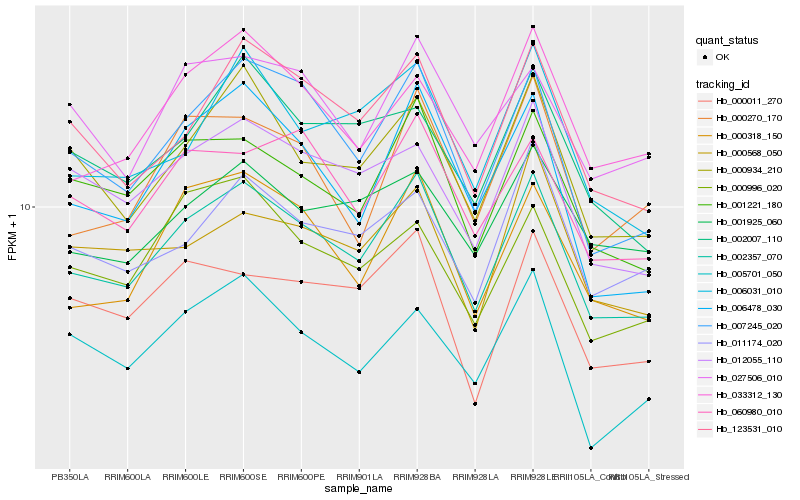
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002357_070 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 2 |
Hb_007245_020 |
0.046028484 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 3 |
Hb_001221_180 |
0.0529345033 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_123531_010 |
0.0562044735 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 5 |
Hb_000934_210 |
0.0569379347 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 6 |
Hb_000318_150 |
0.0569405759 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 7 |
Hb_012055_110 |
0.0618815261 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 8 |
Hb_060980_010 |
0.0622772097 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_006478_030 |
0.0643087583 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 10 |
Hb_011174_020 |
0.0659197241 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 11 |
Hb_033312_130 |
0.0663942345 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006031_010 |
0.0669402206 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 13 |
Hb_000011_270 |
0.0684071079 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 14 |
Hb_002007_110 |
0.0686589945 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 15 |
Hb_027506_010 |
0.0693343688 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 16 |
Hb_000996_020 |
0.0693813626 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 17 |
Hb_001925_060 |
0.0721205392 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 18 |
Hb_005701_050 |
0.0723614063 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 19 |
Hb_000270_170 |
0.0725822473 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 20 |
Hb_000568_050 |
0.0729589396 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |