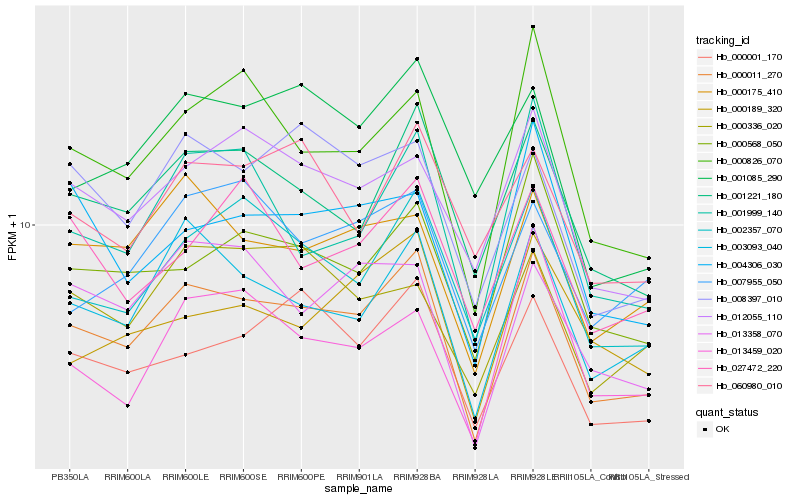
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_270 |
0.0 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 2 |
Hb_003093_040 |
0.0569261589 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_001221_180 |
0.058193718 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_000568_050 |
0.0676342982 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000175_410 |
0.0683598781 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_002357_070 |
0.0684071079 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 7 |
Hb_000001_170 |
0.0703231317 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_012055_110 |
0.0711066511 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 9 |
Hb_000826_070 |
0.0733812043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008397_010 |
0.0774713436 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 11 |
Hb_013358_070 |
0.0798008882 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_001999_140 |
0.0798475279 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_060980_010 |
0.0806178325 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_004306_030 |
0.0829078815 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 15 |
Hb_000189_320 |
0.084310904 |
- |
- |
PREDICTED: multisubstrate pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 16 |
Hb_027472_220 |
0.0844712885 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_007955_050 |
0.0847521733 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_000336_020 |
0.0848903946 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 19 |
Hb_001085_290 |
0.0865807296 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 20 |
Hb_013459_020 |
0.0879565467 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |