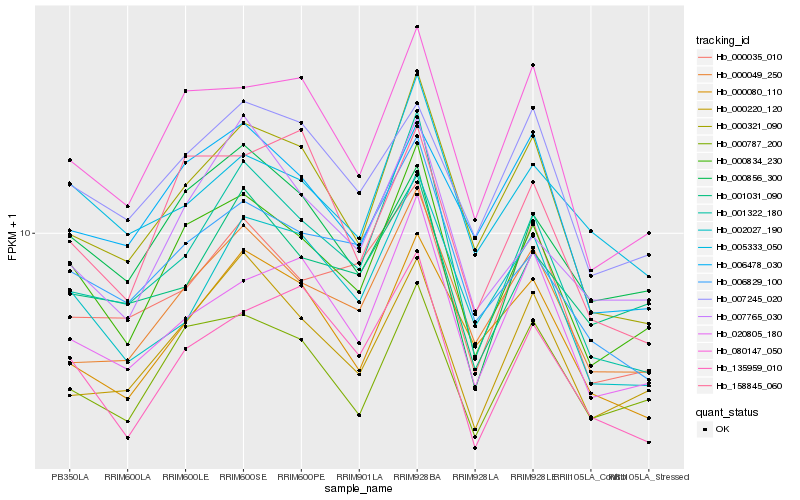
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000834_230 |
0.0 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 2 |
Hb_000787_200 |
0.0693077088 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_080147_050 |
0.0717538248 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 4 |
Hb_000321_090 |
0.0820197358 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 5 |
Hb_000049_250 |
0.0871696251 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000220_120 |
0.0880870257 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001322_180 |
0.0885258727 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 8 |
Hb_006829_100 |
0.0886686372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_020805_180 |
0.088684652 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 10 |
Hb_007765_030 |
0.091442995 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 11 |
Hb_135959_010 |
0.0934453599 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 12 |
Hb_007245_020 |
0.0979778487 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000035_010 |
0.0992475937 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000856_300 |
0.1002034598 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 15 |
Hb_005333_050 |
0.1008454528 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_006478_030 |
0.101373219 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 17 |
Hb_002027_190 |
0.1014255311 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 18 |
Hb_000080_110 |
0.1023216765 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001031_090 |
0.1024786901 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 20 |
Hb_158845_060 |
0.1030394439 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |