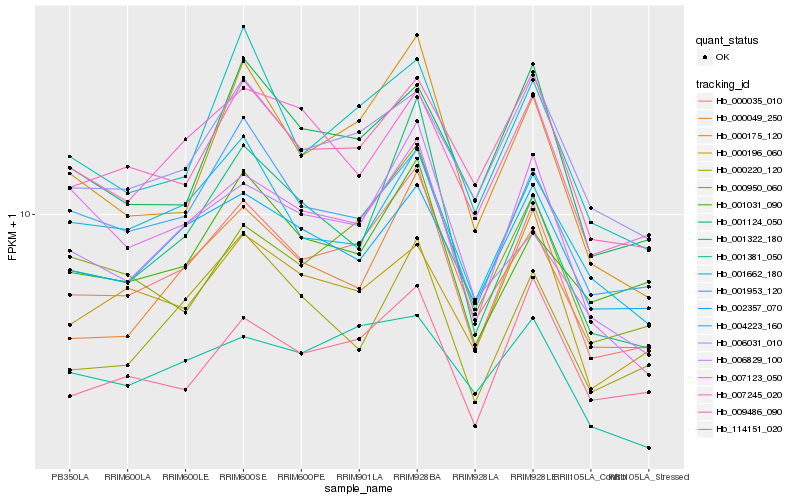
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_010 |
0.0 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000049_250 |
0.0765037483 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000175_120 |
0.0788141831 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 4 |
Hb_009486_090 |
0.0807056604 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_006031_010 |
0.0839554861 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 6 |
Hb_114151_020 |
0.0851648499 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 7 |
Hb_000950_060 |
0.0859312261 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 8 |
Hb_002357_070 |
0.0875511847 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 9 |
Hb_001662_180 |
0.0879527861 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 10 |
Hb_004223_160 |
0.0884459946 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001322_180 |
0.090647695 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 12 |
Hb_001031_090 |
0.0914395865 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 13 |
Hb_006829_100 |
0.0914696233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007245_020 |
0.0933387707 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000220_120 |
0.0947460153 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001124_050 |
0.0950237111 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 17 |
Hb_001953_120 |
0.0954871435 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000196_060 |
0.0958617715 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 19 |
Hb_001381_050 |
0.0960752038 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 20 |
Hb_007123_050 |
0.0964551508 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |