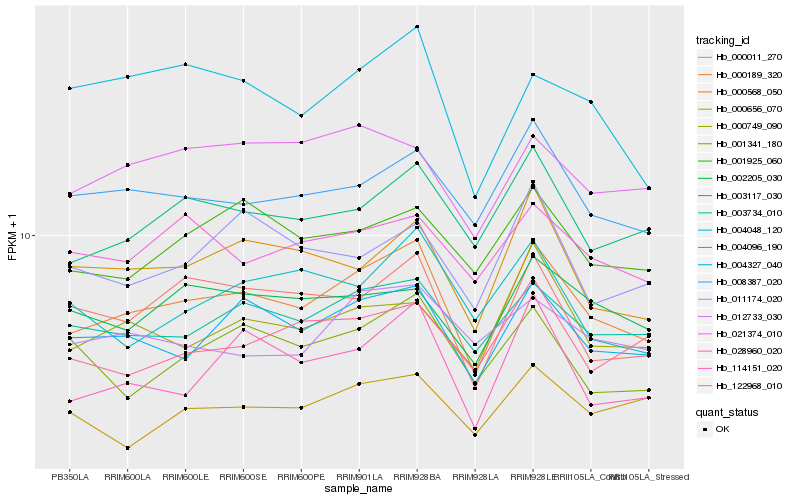
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_320 |
0.0 |
- |
- |
PREDICTED: multisubstrate pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004327_040 |
0.0682626788 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 3 |
Hb_003117_030 |
0.0780084426 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 4 |
Hb_003734_010 |
0.0788001018 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 5 |
Hb_008387_020 |
0.0834209385 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 6 |
Hb_000011_270 |
0.084310904 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 7 |
Hb_000568_050 |
0.0846892096 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_114151_020 |
0.0851719044 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 9 |
Hb_000749_090 |
0.0869927679 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001925_060 |
0.0899354434 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 11 |
Hb_002205_030 |
0.0932593462 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 12 |
Hb_122968_010 |
0.0967552167 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 13 |
Hb_028960_020 |
0.0968759211 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001341_180 |
0.0982043168 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 15 |
Hb_021374_010 |
0.0987488592 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 16 |
Hb_011174_020 |
0.0996402782 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 17 |
Hb_004096_190 |
0.1001005399 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 18 |
Hb_000656_070 |
0.1011060615 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_012733_030 |
0.1011815807 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 20 |
Hb_004048_120 |
0.1012745383 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |