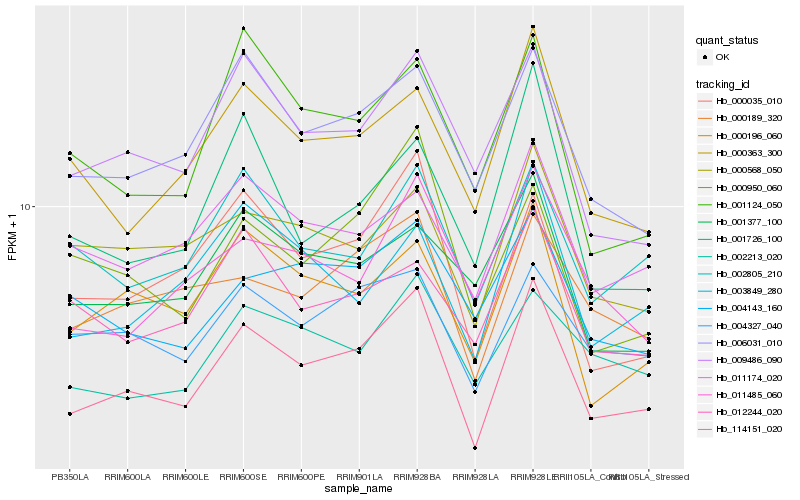
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114151_020 |
0.0 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 2 |
Hb_006031_010 |
0.082465016 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 3 |
Hb_000035_010 |
0.0851648499 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000189_320 |
0.0851719044 |
- |
- |
PREDICTED: multisubstrate pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009486_090 |
0.0862479987 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000196_060 |
0.0875776307 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 7 |
Hb_004327_040 |
0.0882903616 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 8 |
Hb_000363_300 |
0.0902583151 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000568_050 |
0.0931813359 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_011174_020 |
0.0932089698 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 11 |
Hb_002805_210 |
0.0938278481 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 12 |
Hb_003849_280 |
0.0941720221 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 13 |
Hb_011485_060 |
0.0961271737 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 14 |
Hb_000950_060 |
0.0972752851 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 15 |
Hb_002213_020 |
0.0978825441 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004143_160 |
0.0999577946 |
- |
- |
SAB, putative [Ricinus communis] |
| 17 |
Hb_001726_100 |
0.10044071 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001124_050 |
0.1008853753 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 19 |
Hb_012244_020 |
0.102489226 |
- |
- |
calpain, putative [Ricinus communis] |
| 20 |
Hb_001377_100 |
0.103532844 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |