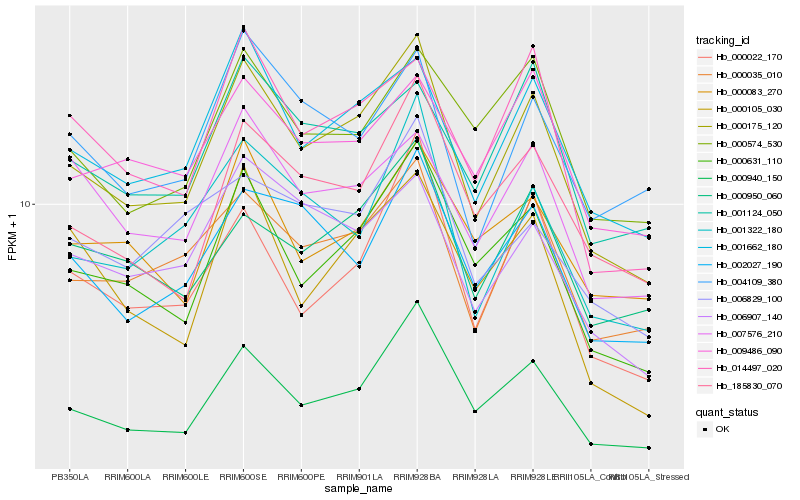
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_120 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 2 |
Hb_000940_150 |
0.066953442 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 3 |
Hb_000631_110 |
0.0712992196 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 4 |
Hb_001662_180 |
0.0735993271 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 5 |
Hb_000035_010 |
0.0788141831 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_185830_070 |
0.0900274788 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_014497_020 |
0.0908122314 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 8 |
Hb_001322_180 |
0.095180641 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 9 |
Hb_000022_170 |
0.0957160668 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 10 |
Hb_002027_190 |
0.095918722 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 11 |
Hb_006907_140 |
0.0978564716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000950_060 |
0.0981569427 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 13 |
Hb_001124_050 |
0.1003028835 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 14 |
Hb_007576_210 |
0.1024046645 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 15 |
Hb_000574_530 |
0.1033685638 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 16 |
Hb_004109_380 |
0.1064641541 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 17 |
Hb_000105_030 |
0.107496702 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 18 |
Hb_009486_090 |
0.1079749721 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000083_270 |
0.1079881797 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 20 |
Hb_006829_100 |
0.1099637316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |