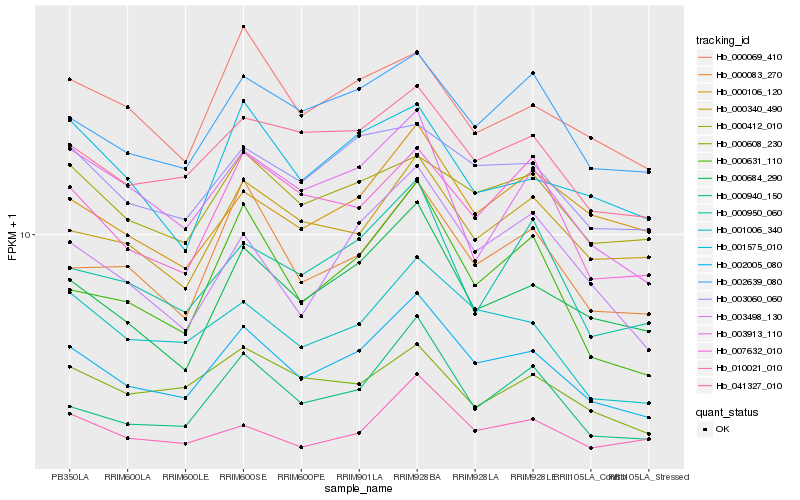
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000684_290 |
0.0753861561 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 3 |
Hb_001006_340 |
0.0805008998 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 4 |
Hb_041327_010 |
0.0860641494 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 5 |
Hb_002639_080 |
0.0876958654 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000083_270 |
0.087914684 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003498_130 |
0.0888189642 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 8 |
Hb_000631_110 |
0.0904654314 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 9 |
Hb_003060_060 |
0.092155626 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 10 |
Hb_007632_010 |
0.094412088 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000340_490 |
0.0962991261 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 12 |
Hb_000412_010 |
0.0986196283 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000940_150 |
0.0998981991 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 14 |
Hb_000950_060 |
0.1003353358 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 15 |
Hb_003913_110 |
0.1012852623 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 16 |
Hb_001575_010 |
0.1036300001 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000608_230 |
0.1049155852 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000106_120 |
0.1052091853 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 19 |
Hb_010021_010 |
0.1059598834 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000069_410 |
0.106211359 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |