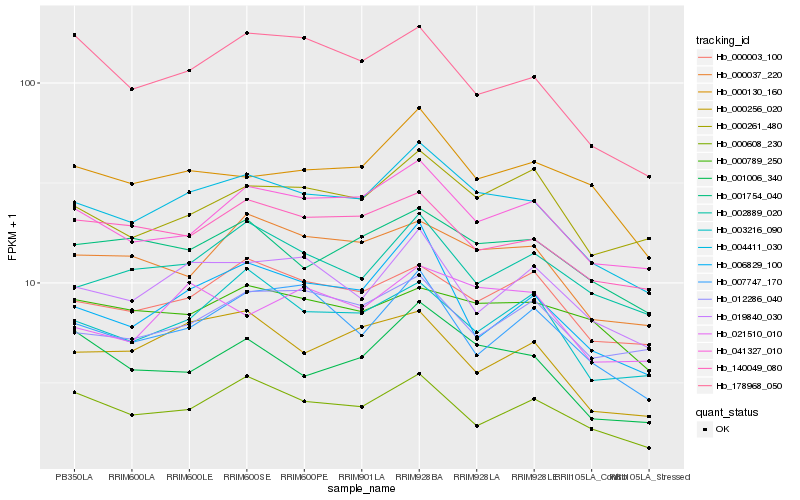
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004411_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 2 |
Hb_041327_010 |
0.0750005963 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 3 |
Hb_000037_220 |
0.0849017325 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 4 |
Hb_001754_040 |
0.0868970287 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 5 |
Hb_140049_080 |
0.087468959 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 6 |
Hb_000003_100 |
0.0875384146 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 7 |
Hb_019840_030 |
0.0878953234 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000130_160 |
0.0879054676 |
- |
- |
PREDICTED: protein DJ-1 homolog B-like [Jatropha curcas] |
| 9 |
Hb_001006_340 |
0.0897246472 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 10 |
Hb_000608_230 |
0.0907043647 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003216_090 |
0.0933440269 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002889_020 |
0.0933734087 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 13 |
Hb_000261_480 |
0.093520424 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 14 |
Hb_006829_100 |
0.0938761554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_021510_010 |
0.0958476878 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 16 |
Hb_178968_050 |
0.0961374421 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 17 |
Hb_000256_020 |
0.0988658397 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 18 |
Hb_007747_170 |
0.1015369714 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 19 |
Hb_012286_040 |
0.1018981218 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 20 |
Hb_000789_250 |
0.1031924068 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |