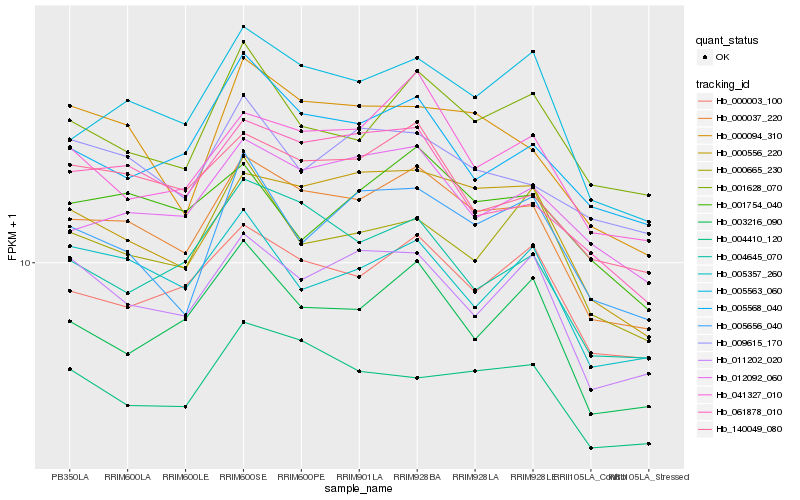
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000037_220 |
0.0 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 2 |
Hb_005563_060 |
0.0558711524 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 3 |
Hb_000556_220 |
0.0567257244 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 4 |
Hb_000003_100 |
0.0604281996 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 5 |
Hb_140049_080 |
0.0644485209 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 6 |
Hb_000094_310 |
0.0679858993 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 7 |
Hb_041327_010 |
0.0701289807 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 8 |
Hb_011202_020 |
0.0706745212 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_012092_060 |
0.0708613003 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 10 |
Hb_004410_120 |
0.071996576 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 11 |
Hb_061878_010 |
0.0720095948 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 12 |
Hb_004645_070 |
0.0729505744 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 13 |
Hb_003216_090 |
0.0743130702 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001628_070 |
0.076497117 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_005568_040 |
0.0774152714 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005656_040 |
0.0801785172 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 17 |
Hb_005357_260 |
0.0813008813 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 18 |
Hb_000665_230 |
0.0830368328 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 19 |
Hb_009615_170 |
0.0834494778 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001754_040 |
0.0841663642 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |