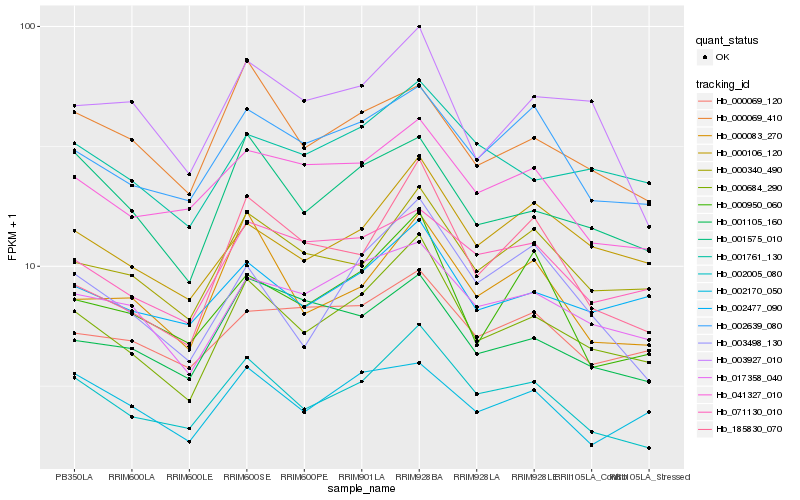
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_290 |
0.0 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 2 |
Hb_002005_080 |
0.0753861561 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 3 |
Hb_017358_040 |
0.0851209735 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 4 |
Hb_000340_490 |
0.0890350543 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 5 |
Hb_000106_120 |
0.0891830833 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 6 |
Hb_002477_090 |
0.0901689866 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001761_130 |
0.0906588255 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001575_010 |
0.0915463452 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_002639_080 |
0.0978091217 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_185830_070 |
0.1011274659 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_071130_010 |
0.1016633851 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000083_270 |
0.1036314732 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001105_160 |
0.1051701452 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 14 |
Hb_000950_060 |
0.1056353589 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 15 |
Hb_000069_120 |
0.1067817013 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 16 |
Hb_003927_010 |
0.1069688079 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_003498_130 |
0.1074929548 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 18 |
Hb_041327_010 |
0.1083106932 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 19 |
Hb_000069_410 |
0.1100670914 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 20 |
Hb_002170_050 |
0.1102959899 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09060 [Jatropha curcas] |