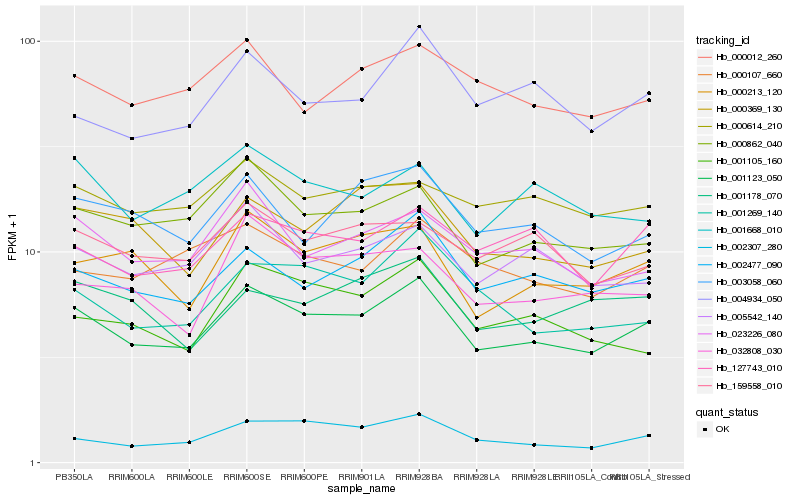
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002477_090 |
0.0738657174 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000862_040 |
0.0827677924 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 4 |
Hb_000213_120 |
0.0859163725 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_023226_080 |
0.0869079081 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005542_140 |
0.0872079001 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 7 |
Hb_127743_010 |
0.0882688103 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000012_260 |
0.0900965146 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 9 |
Hb_003058_060 |
0.0903461189 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 10 |
Hb_002307_280 |
0.0904126928 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_159558_010 |
0.0904454329 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 12 |
Hb_000369_130 |
0.0915881962 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 13 |
Hb_000614_210 |
0.0931063686 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000107_660 |
0.0935006025 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 15 |
Hb_004934_050 |
0.0939095603 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 16 |
Hb_001668_010 |
0.0951740807 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 17 |
Hb_001178_070 |
0.0953783223 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 18 |
Hb_001269_140 |
0.0955566188 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 19 |
Hb_032808_030 |
0.0961370683 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001105_160 |
0.0975415306 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |