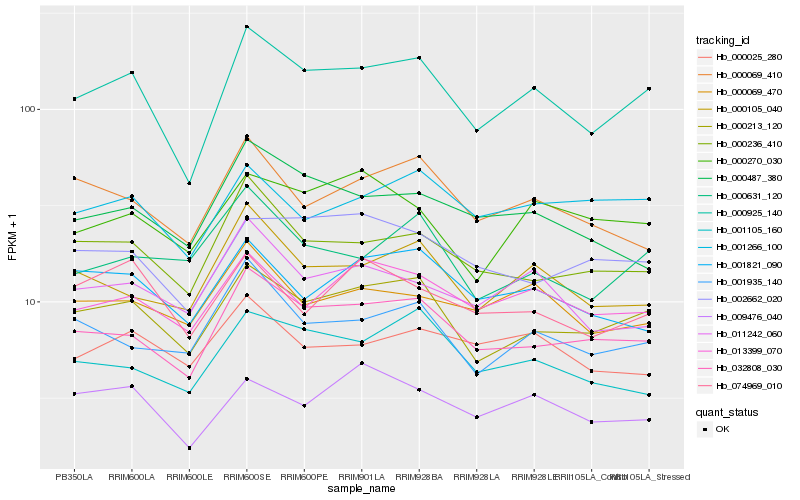
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000925_140 |
0.0 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 2 |
Hb_000213_120 |
0.0722730227 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_032808_030 |
0.0788992382 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000236_410 |
0.1001030479 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 5 |
Hb_000105_040 |
0.1045566656 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 6 |
Hb_013399_070 |
0.1055625611 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001935_140 |
0.1087165958 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 8 |
Hb_001266_100 |
0.1111547223 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 9 |
Hb_011242_060 |
0.1125714495 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 10 |
Hb_009476_040 |
0.112693905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001105_160 |
0.1132591036 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 12 |
Hb_000069_470 |
0.1133350102 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002662_020 |
0.1139730695 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 14 |
Hb_000069_410 |
0.1141131134 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 15 |
Hb_000270_030 |
0.1160867623 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 16 |
Hb_001821_090 |
0.1168500783 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 17 |
Hb_000487_380 |
0.1173944697 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000631_120 |
0.118953469 |
- |
- |
hypothetical protein CISIN_1g0431902mg, partial [Citrus sinensis] |
| 19 |
Hb_000025_280 |
0.1194309613 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 20 |
Hb_074969_010 |
0.1198114094 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |