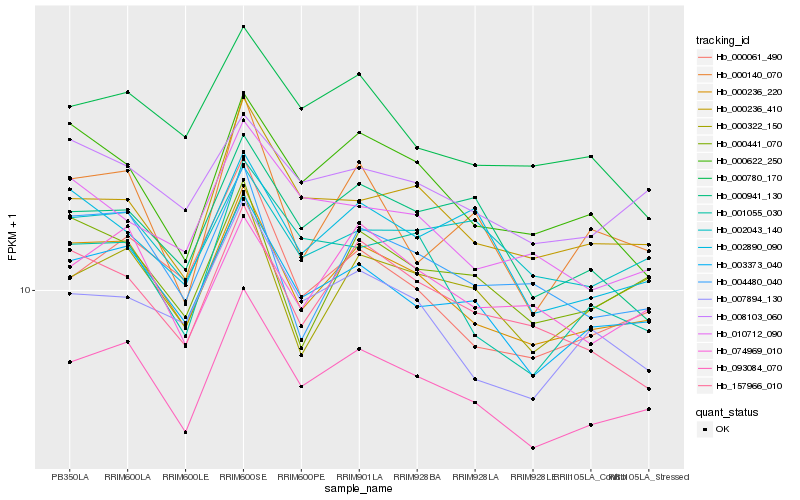
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093084_070 |
0.0 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 2 |
Hb_000236_220 |
0.0417398127 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 3 |
Hb_003373_040 |
0.0436315272 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 4 |
Hb_000236_410 |
0.0706420103 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 5 |
Hb_001055_030 |
0.0816381512 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000061_490 |
0.0835597945 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 7 |
Hb_074969_010 |
0.0846362978 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 8 |
Hb_008103_060 |
0.0851451641 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_000322_150 |
0.0859478265 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007894_130 |
0.086996332 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 11 |
Hb_002890_090 |
0.0876664462 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 12 |
Hb_000140_070 |
0.0880711558 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002043_140 |
0.0884786001 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 14 |
Hb_000622_250 |
0.0888251293 |
- |
- |
atpob1, putative [Ricinus communis] |
| 15 |
Hb_000941_130 |
0.0894185483 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 16 |
Hb_000780_170 |
0.0903887 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 17 |
Hb_157966_010 |
0.0919318622 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 18 |
Hb_010712_090 |
0.092336083 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 19 |
Hb_000441_070 |
0.0926035069 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004480_040 |
0.0927627974 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |