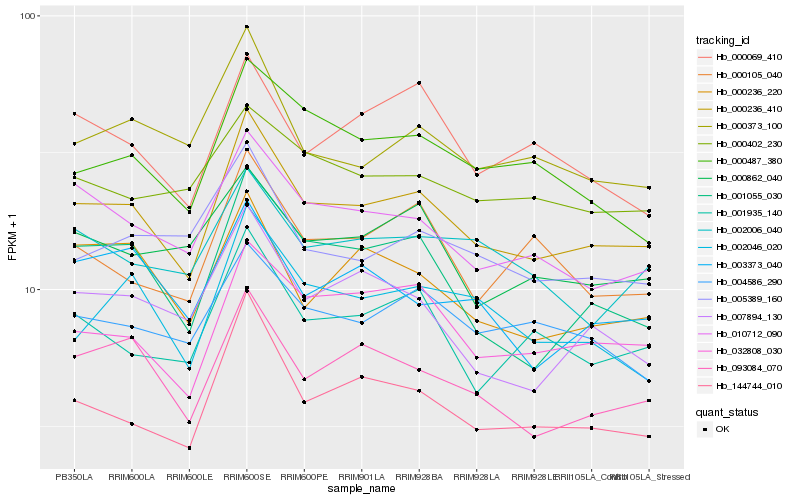
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_410 |
0.0 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 2 |
Hb_032808_030 |
0.0667945032 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_144744_010 |
0.0702363618 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 4 |
Hb_093084_070 |
0.0706420103 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 5 |
Hb_010712_090 |
0.0708738934 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 6 |
Hb_001055_030 |
0.074970592 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001935_140 |
0.0798590051 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 8 |
Hb_000236_220 |
0.0832466082 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 9 |
Hb_004586_290 |
0.0834635299 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 10 |
Hb_000373_100 |
0.084183157 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 11 |
Hb_000487_380 |
0.0861084922 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002046_020 |
0.0863111667 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 13 |
Hb_000862_040 |
0.0884780607 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 14 |
Hb_007894_130 |
0.0887289083 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 15 |
Hb_003373_040 |
0.0891119412 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 16 |
Hb_005389_160 |
0.0892870581 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 17 |
Hb_002006_040 |
0.0896464872 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 18 |
Hb_000069_410 |
0.0896791469 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 19 |
Hb_000105_040 |
0.0896875194 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 20 |
Hb_000402_230 |
0.0901966849 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |