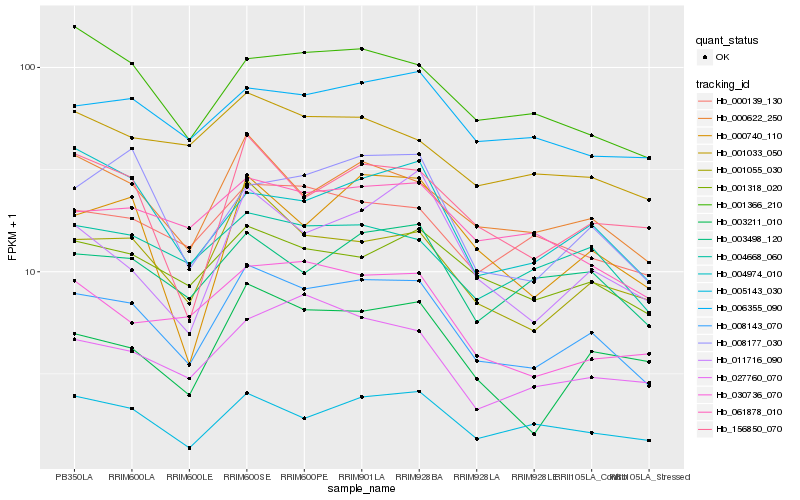
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008143_070 |
0.0 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 2 |
Hb_001055_030 |
0.0845970056 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000622_250 |
0.0856893097 |
- |
- |
atpob1, putative [Ricinus communis] |
| 4 |
Hb_005143_030 |
0.0966898181 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 5 |
Hb_003498_120 |
0.1046945497 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 6 |
Hb_001318_020 |
0.1047421699 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_004668_060 |
0.1056199156 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000139_130 |
0.1065545267 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 9 |
Hb_008177_030 |
0.1068497544 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_030736_070 |
0.1076836835 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_061878_010 |
0.1080148012 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 12 |
Hb_003211_010 |
0.1085503065 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 13 |
Hb_001366_210 |
0.1088352238 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 14 |
Hb_000740_110 |
0.1089234416 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 15 |
Hb_027760_070 |
0.1094277035 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 16 |
Hb_156850_070 |
0.1112312133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006355_090 |
0.1118259917 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 18 |
Hb_004974_010 |
0.1127363414 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 19 |
Hb_001033_050 |
0.1133395309 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 20 |
Hb_011716_090 |
0.1134730728 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |