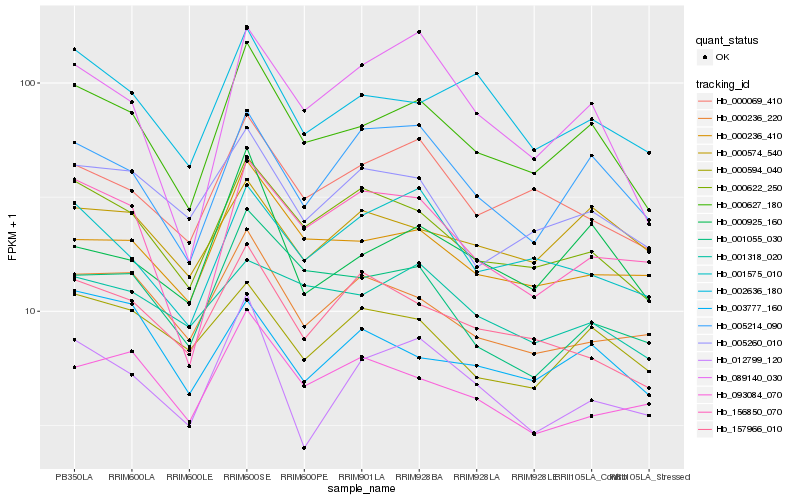
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000627_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000622_250 |
0.0776542754 |
- |
- |
atpob1, putative [Ricinus communis] |
| 3 |
Hb_000236_410 |
0.0922099335 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 4 |
Hb_000069_410 |
0.0955067364 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 5 |
Hb_002636_180 |
0.0965457283 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 6 |
Hb_005214_090 |
0.0978069881 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 7 |
Hb_000925_160 |
0.097840025 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005260_010 |
0.1012723701 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001055_030 |
0.1025404079 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_156850_070 |
0.1028242086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000236_220 |
0.1035881486 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_157966_010 |
0.1036801002 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 13 |
Hb_001575_010 |
0.1043209089 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000594_040 |
0.1066000763 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 15 |
Hb_089140_030 |
0.109564507 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 16 |
Hb_012799_120 |
0.1105387439 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001318_020 |
0.1107387631 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 18 |
Hb_093084_070 |
0.1112368769 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_000574_540 |
0.1115359394 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_003777_160 |
0.1117586884 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |