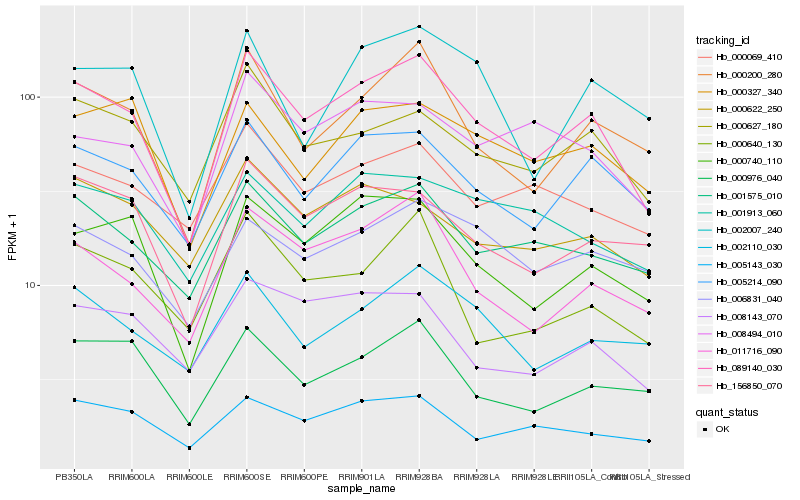
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089140_030 |
0.0 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 2 |
Hb_000200_280 |
0.0966815785 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 3 |
Hb_011716_090 |
0.098382258 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 4 |
Hb_000740_110 |
0.0986346589 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 5 |
Hb_005214_090 |
0.0992374467 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 6 |
Hb_001575_010 |
0.1032456848 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000627_180 |
0.109564507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_156850_070 |
0.113660862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008494_010 |
0.1201909634 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 10 |
Hb_005143_030 |
0.1204266452 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 11 |
Hb_000976_040 |
0.1205941768 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 12 |
Hb_002110_030 |
0.1206122638 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 13 |
Hb_008143_070 |
0.1210847784 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 14 |
Hb_000069_410 |
0.1245412814 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 15 |
Hb_000327_340 |
0.1249015543 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 16 |
Hb_000622_250 |
0.1251024761 |
- |
- |
atpob1, putative [Ricinus communis] |
| 17 |
Hb_001913_060 |
0.125307353 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 18 |
Hb_000640_130 |
0.1258286842 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_006831_040 |
0.1263662757 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002007_240 |
0.1264513619 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |