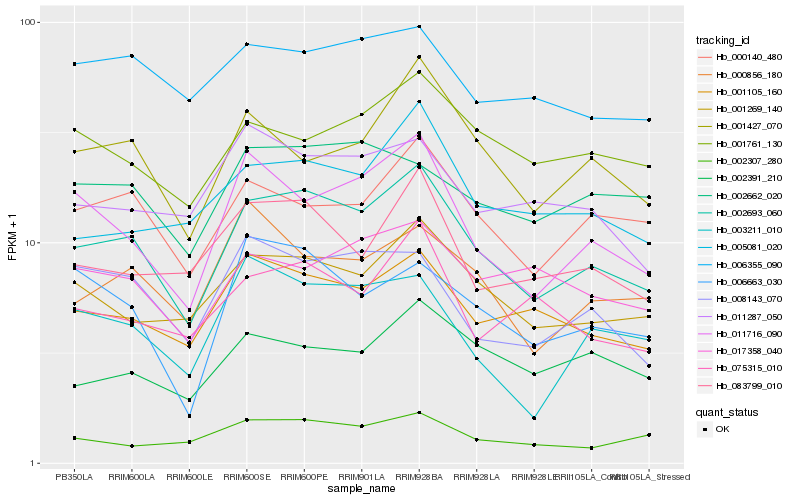
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002693_060 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001269_140 |
0.0989217304 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 3 |
Hb_001105_160 |
0.101569547 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 4 |
Hb_011716_090 |
0.1033358896 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 5 |
Hb_000140_480 |
0.1063189221 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 6 |
Hb_001427_070 |
0.1110869124 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 7 |
Hb_083799_010 |
0.1128078052 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_005081_020 |
0.1173212788 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 9 |
Hb_002391_210 |
0.1194024262 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 10 |
Hb_000856_180 |
0.1215493305 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 11 |
Hb_001761_130 |
0.1219631786 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008143_070 |
0.123110477 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 13 |
Hb_006355_090 |
0.1234042742 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 14 |
Hb_002307_280 |
0.1240380855 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 15 |
Hb_075315_010 |
0.1244493628 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 16 |
Hb_002662_020 |
0.1251182099 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 17 |
Hb_011287_050 |
0.1252542385 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 18 |
Hb_006663_030 |
0.126086203 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 19 |
Hb_003211_010 |
0.1263487013 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 20 |
Hb_017358_040 |
0.126386759 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |