| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
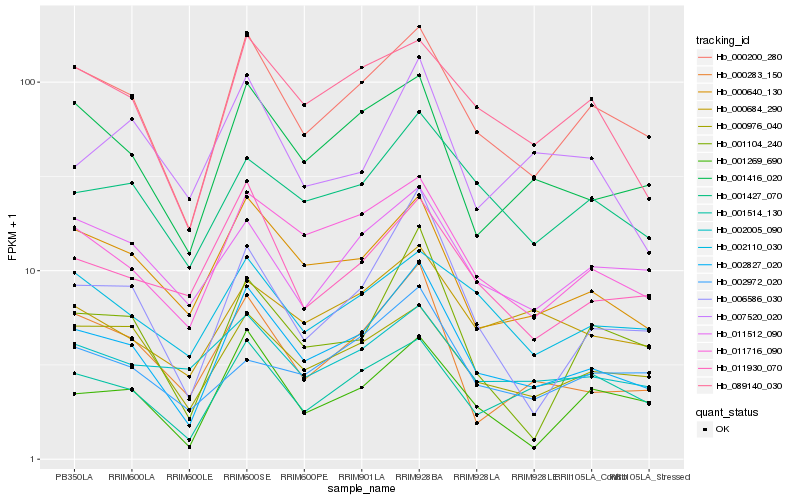
Hb_002827_020 |
0.0 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 2 |
Hb_000200_280 |
0.0863068847 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 3 |
Hb_011716_090 |
0.1072437606 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 4 |
Hb_000640_130 |
0.1125168975 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_006586_030 |
0.1152271404 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 6 |
Hb_001416_020 |
0.1181211104 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 7 |
Hb_000283_150 |
0.1207025968 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 8 |
Hb_089140_030 |
0.1264661191 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 9 |
Hb_000976_040 |
0.1305725839 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 10 |
Hb_001427_070 |
0.1320364025 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 11 |
Hb_001269_690 |
0.1364065016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001104_240 |
0.1400626899 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_001514_130 |
0.140398853 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 14 |
Hb_011930_070 |
0.1418885233 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 15 |
Hb_000684_290 |
0.1440952037 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 16 |
Hb_011512_090 |
0.1485389841 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 17 |
Hb_002110_030 |
0.1495985561 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002005_090 |
0.1507548039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002972_020 |
0.1533034416 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 20 |
Hb_007520_020 |
0.1533837107 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
1-deoxy-D-xylulose-5-phosphate reductoisomerase [Hevea brasiliensis] |