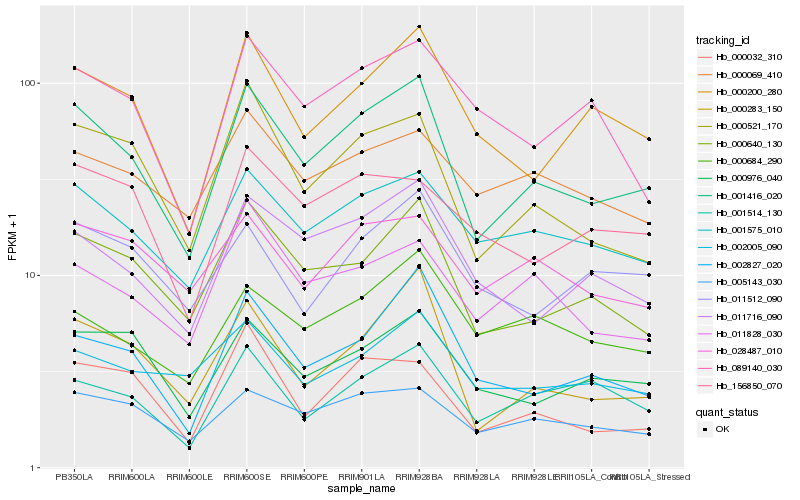
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001416_020 |
0.0 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 2 |
Hb_000640_130 |
0.0992537801 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000521_170 |
0.1067680214 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 4 |
Hb_000283_150 |
0.1072150874 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 5 |
Hb_001575_010 |
0.1113256589 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_005143_030 |
0.1124363372 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 7 |
Hb_011716_090 |
0.1127344705 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 8 |
Hb_000200_280 |
0.1130030851 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 9 |
Hb_000976_040 |
0.1170000854 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 10 |
Hb_002827_020 |
0.1181211104 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 11 |
Hb_000032_310 |
0.127721977 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_001514_130 |
0.1317615093 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 13 |
Hb_011828_030 |
0.1334581375 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 14 |
Hb_028487_010 |
0.1351383222 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 15 |
Hb_156850_070 |
0.1358927179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002005_090 |
0.1362081395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000684_290 |
0.1373865021 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 18 |
Hb_011512_090 |
0.1375838049 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 19 |
Hb_089140_030 |
0.1394972153 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 20 |
Hb_000069_410 |
0.1396171825 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |