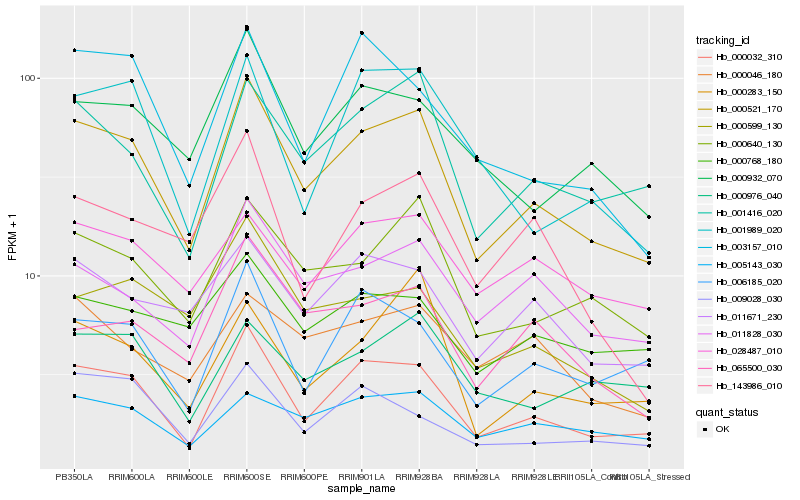
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_170 |
0.0 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 2 |
Hb_000032_310 |
0.0607875414 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_000640_130 |
0.1059568728 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001416_020 |
0.1067680214 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 5 |
Hb_000932_070 |
0.1257993285 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 6 |
Hb_011828_030 |
0.1261243623 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 7 |
Hb_011671_230 |
0.1273629489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006185_020 |
0.1293360178 |
- |
- |
PREDICTED: midasin [Jatropha curcas] |
| 9 |
Hb_065500_030 |
0.134573116 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 10 |
Hb_000599_130 |
0.1360184684 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 11 |
Hb_000283_150 |
0.1379344886 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 12 |
Hb_005143_030 |
0.1383099831 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 13 |
Hb_000976_040 |
0.1383309175 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 14 |
Hb_001989_020 |
0.1391347911 |
- |
- |
hypothetical protein JCGZ_15521 [Jatropha curcas] |
| 15 |
Hb_003157_010 |
0.1402533075 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 16 |
Hb_009028_030 |
0.1403700947 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 17 |
Hb_000046_180 |
0.1411642479 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 18 |
Hb_028487_010 |
0.1426228882 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000768_180 |
0.1428135089 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 20 |
Hb_143986_010 |
0.144269712 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |