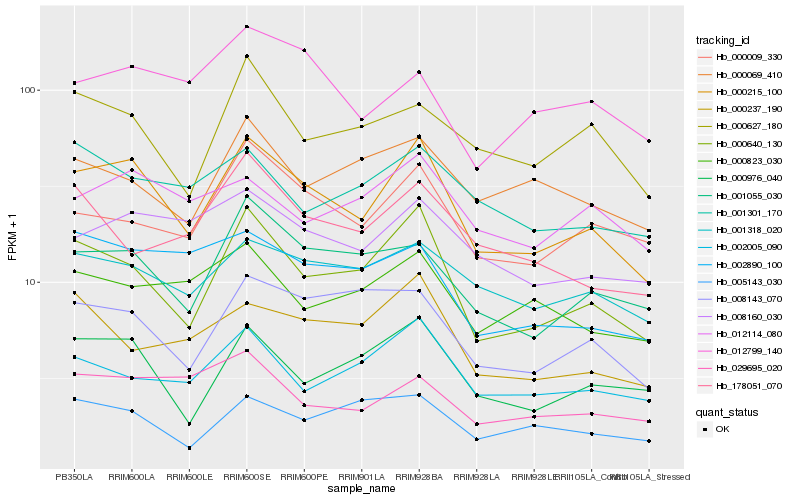
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000640_130 |
0.0848822906 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_001055_030 |
0.1169435192 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000976_040 |
0.1177156448 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 5 |
Hb_000009_330 |
0.1226281809 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_001318_020 |
0.1232029512 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_008160_030 |
0.1246187553 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_008143_070 |
0.1253426375 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 9 |
Hb_002890_100 |
0.131619796 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000627_180 |
0.1321166055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002005_090 |
0.132916597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000237_190 |
0.1369646039 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_029695_020 |
0.1370156708 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000823_030 |
0.1376159257 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 15 |
Hb_012114_080 |
0.1377709768 |
- |
- |
gamma-glutamylcysteine synthetase [Hevea brasiliensis] |
| 16 |
Hb_012799_140 |
0.1403848705 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 17 |
Hb_001301_170 |
0.1406647255 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 18 |
Hb_000069_410 |
0.1419452679 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 19 |
Hb_005143_030 |
0.1419792882 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 20 |
Hb_178051_070 |
0.1436947485 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |