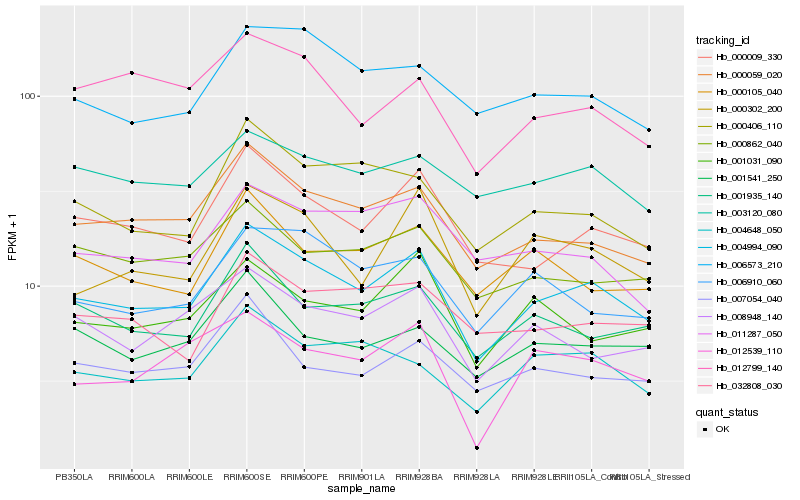
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000009_330 |
0.0785348109 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_000059_020 |
0.0790345403 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 4 |
Hb_001935_140 |
0.095969041 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 5 |
Hb_000302_200 |
0.0971083999 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 6 |
Hb_000406_110 |
0.0995285355 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 7 |
Hb_008948_140 |
0.100550043 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 8 |
Hb_004648_050 |
0.1017465398 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000105_040 |
0.1025708723 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 10 |
Hb_006910_060 |
0.1031096004 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 11 |
Hb_000862_040 |
0.1034797777 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 12 |
Hb_006573_210 |
0.1034986086 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 13 |
Hb_001541_250 |
0.1040229393 |
- |
- |
PREDICTED: integrator complex subunit 9 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_001031_090 |
0.1046307665 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 15 |
Hb_007054_040 |
0.1049611962 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 16 |
Hb_032808_030 |
0.1074681356 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_003120_080 |
0.1078699548 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012539_110 |
0.1093115997 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_011287_050 |
0.1097242409 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 20 |
Hb_012799_140 |
0.1107207294 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |