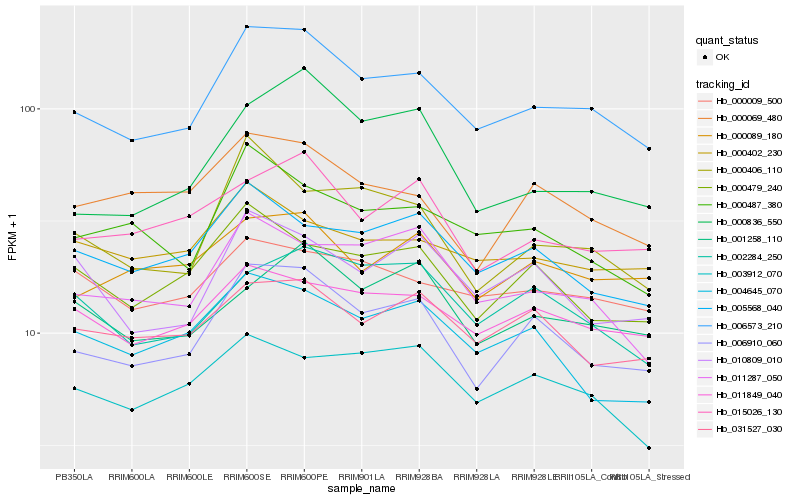
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006573_210 |
0.0 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 2 |
Hb_006910_060 |
0.0600166644 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 3 |
Hb_011849_040 |
0.0826627141 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_011287_050 |
0.0869183923 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 5 |
Hb_000487_380 |
0.0872526164 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000406_110 |
0.0877052409 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 7 |
Hb_015026_130 |
0.0919652042 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 8 |
Hb_031527_030 |
0.0926206133 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_005568_040 |
0.0926398228 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000009_500 |
0.0926806516 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 11 |
Hb_002284_250 |
0.0933059919 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 12 |
Hb_000479_240 |
0.0937329897 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010809_010 |
0.0948338898 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 14 |
Hb_003912_070 |
0.0949980916 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 15 |
Hb_000089_180 |
0.0951803484 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 16 |
Hb_000836_550 |
0.0955581699 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 17 |
Hb_001258_110 |
0.096510314 |
- |
- |
transporter, putative [Ricinus communis] |
| 18 |
Hb_000069_480 |
0.0971843512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000402_230 |
0.0977333414 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_004645_070 |
0.0982976688 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |