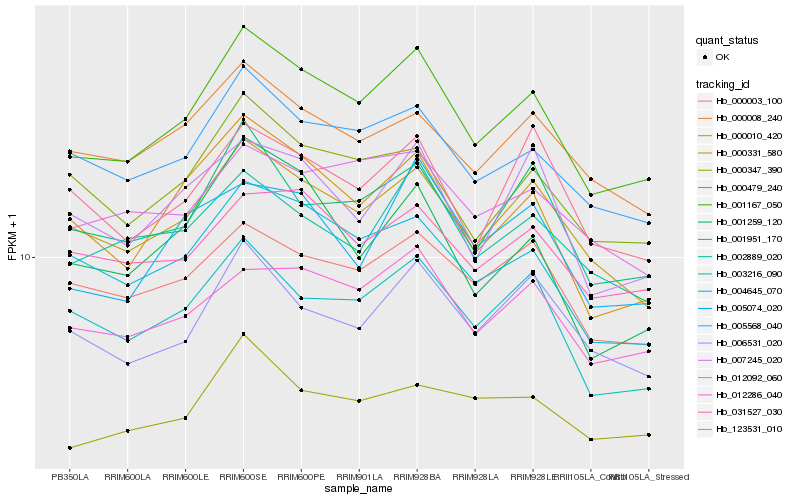
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001167_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 2 |
Hb_000331_580 |
0.05763443 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 3 |
Hb_005568_040 |
0.0619087238 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000010_420 |
0.0661369543 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 5 |
Hb_004645_070 |
0.066404521 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 6 |
Hb_012286_040 |
0.0674819727 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 7 |
Hb_001951_170 |
0.0676198759 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 8 |
Hb_003216_090 |
0.0694225047 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000479_240 |
0.0727844757 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000003_100 |
0.0737979072 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 11 |
Hb_006531_020 |
0.0757723567 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 12 |
Hb_002889_020 |
0.0770008083 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 13 |
Hb_031527_030 |
0.0777415885 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 14 |
Hb_000008_240 |
0.07779872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000347_390 |
0.0791739182 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 16 |
Hb_007245_020 |
0.0796594906 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 17 |
Hb_005074_020 |
0.0807388068 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012092_060 |
0.0808102404 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 19 |
Hb_001259_120 |
0.0828533303 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 20 |
Hb_123531_010 |
0.083599534 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |