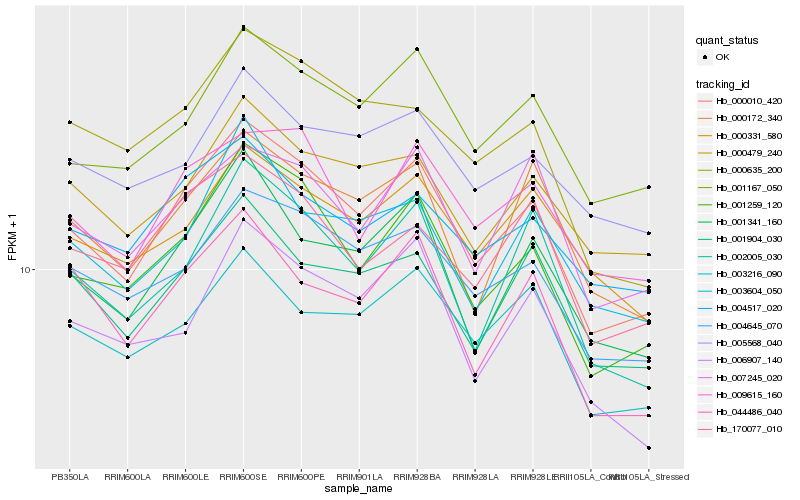
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_420 |
0.0 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 2 |
Hb_001259_120 |
0.0506738304 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 3 |
Hb_004645_070 |
0.062083794 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 4 |
Hb_001167_050 |
0.0661369543 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 5 |
Hb_001341_160 |
0.0670099228 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001904_030 |
0.0723542517 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 7 |
Hb_009615_160 |
0.0740890848 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 8 |
Hb_002005_030 |
0.0746683082 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 9 |
Hb_000172_340 |
0.0757375675 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 10 |
Hb_000479_240 |
0.0765580232 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003216_090 |
0.0785154947 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 12 |
Hb_170077_010 |
0.0786554031 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007245_020 |
0.079067974 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 14 |
Hb_004517_020 |
0.0798520502 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000331_580 |
0.080182945 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 16 |
Hb_044486_040 |
0.0812761398 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 17 |
Hb_000635_200 |
0.0816630134 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 18 |
Hb_003604_050 |
0.0824924618 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_006907_140 |
0.0829154451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005568_040 |
0.0837829119 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |