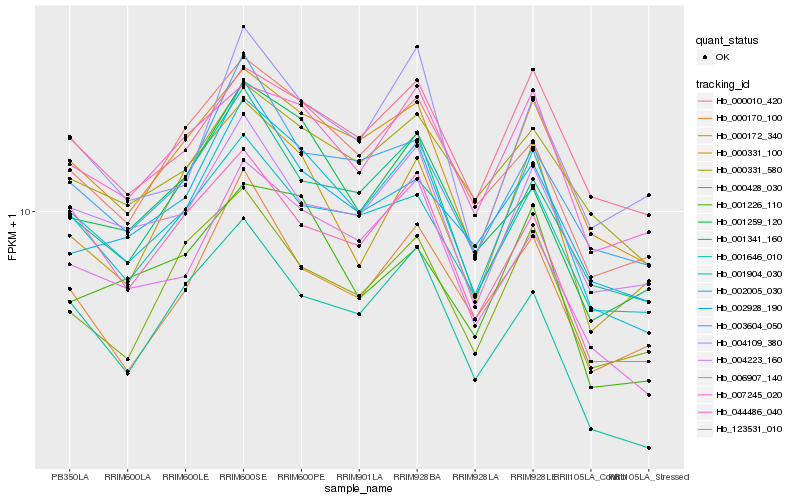
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 2 |
Hb_000172_340 |
0.0578141849 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 3 |
Hb_006907_140 |
0.0603576386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001341_160 |
0.0729799163 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000010_420 |
0.0746683082 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 6 |
Hb_001904_030 |
0.0788791759 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 7 |
Hb_007245_020 |
0.0791615029 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 8 |
Hb_003604_050 |
0.0795178009 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001259_120 |
0.080708033 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 10 |
Hb_004223_160 |
0.0826116952 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_044486_040 |
0.0852096976 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 12 |
Hb_004109_380 |
0.0858658256 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 13 |
Hb_000170_100 |
0.0865851306 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 14 |
Hb_000331_100 |
0.0867481433 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001646_010 |
0.0871674488 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 16 |
Hb_123531_010 |
0.0871838331 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 17 |
Hb_000428_030 |
0.0888228184 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 18 |
Hb_000331_580 |
0.089364642 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 19 |
Hb_001226_110 |
0.0901785539 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 20 |
Hb_002928_190 |
0.0915960153 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |