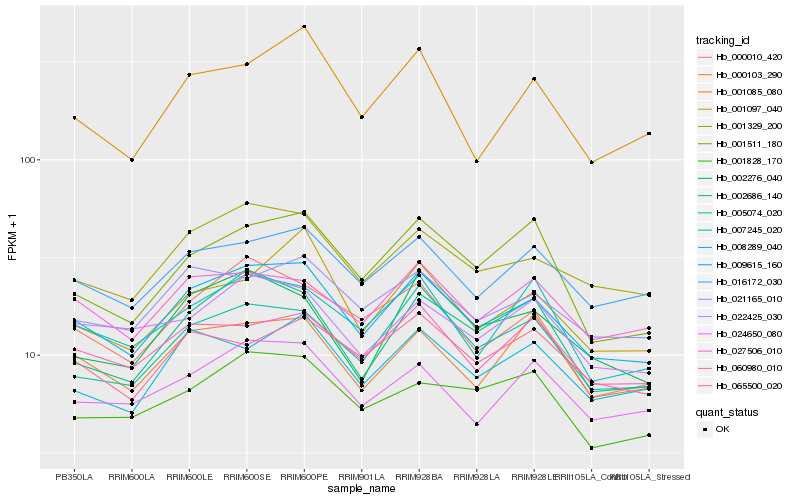
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009615_160 |
0.0 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 2 |
Hb_001329_200 |
0.0522632289 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 3 |
Hb_016172_030 |
0.0628535657 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 4 |
Hb_021165_010 |
0.0672689598 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005074_020 |
0.0686902212 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 6 |
Hb_060980_010 |
0.0734796435 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_024650_080 |
0.0735612603 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 8 |
Hb_000010_420 |
0.0740890848 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 9 |
Hb_027506_010 |
0.0750238276 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 10 |
Hb_002686_140 |
0.0770070616 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008289_040 |
0.0776296926 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 12 |
Hb_007245_020 |
0.0778916611 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 13 |
Hb_022425_030 |
0.0781217887 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 14 |
Hb_000103_290 |
0.078444218 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 15 |
Hb_001097_040 |
0.0793537286 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 16 |
Hb_002276_040 |
0.0806940034 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 17 |
Hb_001828_170 |
0.0810976258 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 18 |
Hb_001085_080 |
0.0824606629 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 19 |
Hb_065500_020 |
0.0829749423 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 20 |
Hb_001511_180 |
0.0831772809 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |