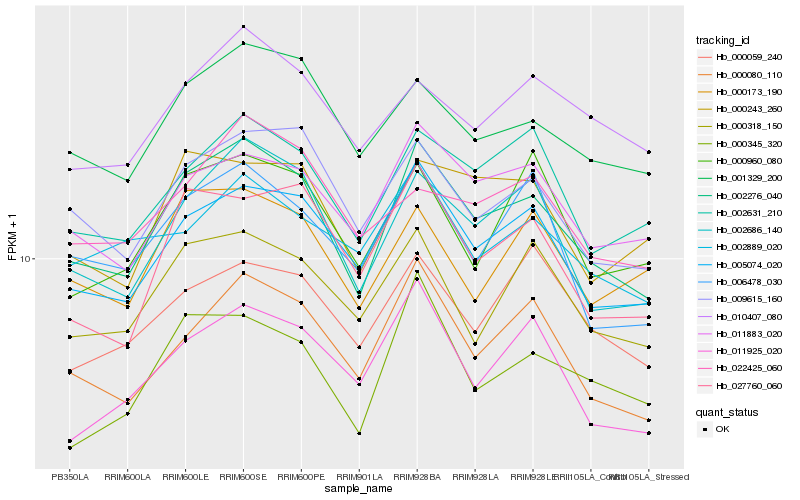
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_040 |
0.0 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 2 |
Hb_002686_140 |
0.0686384029 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005074_020 |
0.0699167945 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000080_110 |
0.0777535861 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009615_160 |
0.0806940034 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 6 |
Hb_011883_020 |
0.082859644 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 7 |
Hb_001329_200 |
0.0842855341 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 8 |
Hb_002631_210 |
0.0856970758 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 9 |
Hb_000318_150 |
0.0897559904 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 10 |
Hb_027760_060 |
0.0910003184 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_006478_030 |
0.0931158216 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 12 |
Hb_000243_260 |
0.0945638301 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000345_320 |
0.098049778 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein ALWAYS EARLY 2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_010407_080 |
0.0988903617 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000173_190 |
0.1001476373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000059_240 |
0.1014140251 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 17 |
Hb_011925_020 |
0.1014509802 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_000960_080 |
0.1020751048 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_022425_060 |
0.1036567607 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_002889_020 |
0.1038523661 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |