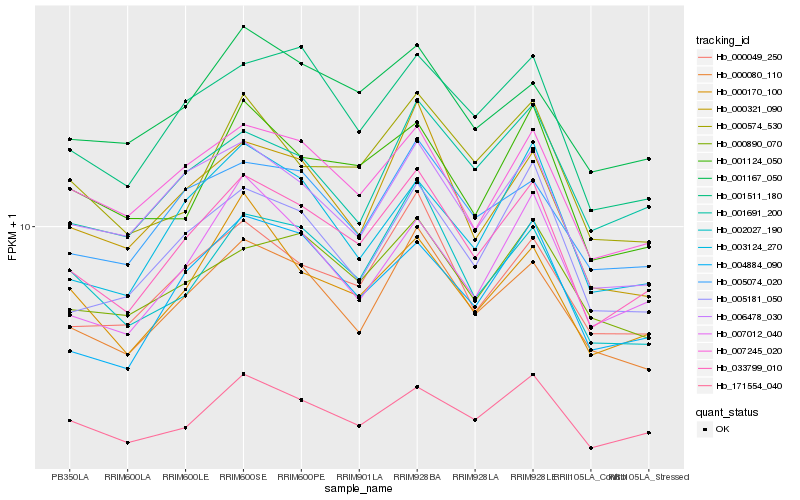
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033799_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 2 |
Hb_000080_110 |
0.0659559702 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_171554_040 |
0.0669825422 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 4 |
Hb_001511_180 |
0.0719472101 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 5 |
Hb_000049_250 |
0.0729133508 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007245_020 |
0.0748725821 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 7 |
Hb_005074_020 |
0.0759674421 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004884_090 |
0.0797669376 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 9 |
Hb_002027_190 |
0.0802666148 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 10 |
Hb_006478_030 |
0.0803920467 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 11 |
Hb_005181_050 |
0.0822764734 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 12 |
Hb_000321_090 |
0.0835747439 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 13 |
Hb_000890_070 |
0.0843921926 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000574_530 |
0.0847584574 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 15 |
Hb_001167_050 |
0.0850622553 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 16 |
Hb_001124_050 |
0.085402394 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 17 |
Hb_001691_200 |
0.0873114396 |
- |
- |
DNA-directed RNA polymerase II subunit RPB2 [Auxenochlorella protothecoides] |
| 18 |
Hb_000170_100 |
0.0874541859 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 19 |
Hb_003124_270 |
0.087504444 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 20 |
Hb_007012_040 |
0.0878991667 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |