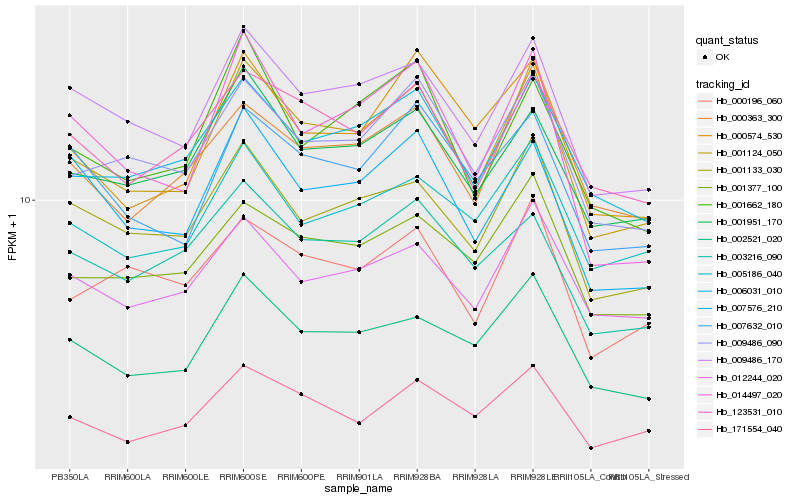
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_050 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 2 |
Hb_009486_170 |
0.0592398002 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 3 |
Hb_009486_090 |
0.0605415903 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_002521_020 |
0.0612642674 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_006031_010 |
0.0661574827 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 6 |
Hb_001133_030 |
0.0672807185 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 7 |
Hb_001951_170 |
0.0680877055 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 8 |
Hb_000574_530 |
0.0711557359 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 9 |
Hb_014497_020 |
0.0712677421 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 10 |
Hb_123531_010 |
0.0719603307 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 11 |
Hb_001377_100 |
0.0723117679 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 12 |
Hb_171554_040 |
0.0724037501 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |
| 13 |
Hb_001662_180 |
0.074437211 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 14 |
Hb_007632_010 |
0.0755110148 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005186_040 |
0.0768265401 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007576_210 |
0.0785055768 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 17 |
Hb_000363_300 |
0.0789884037 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012244_020 |
0.0792158186 |
- |
- |
calpain, putative [Ricinus communis] |
| 19 |
Hb_000196_060 |
0.0807734502 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 20 |
Hb_003216_090 |
0.0820944391 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |