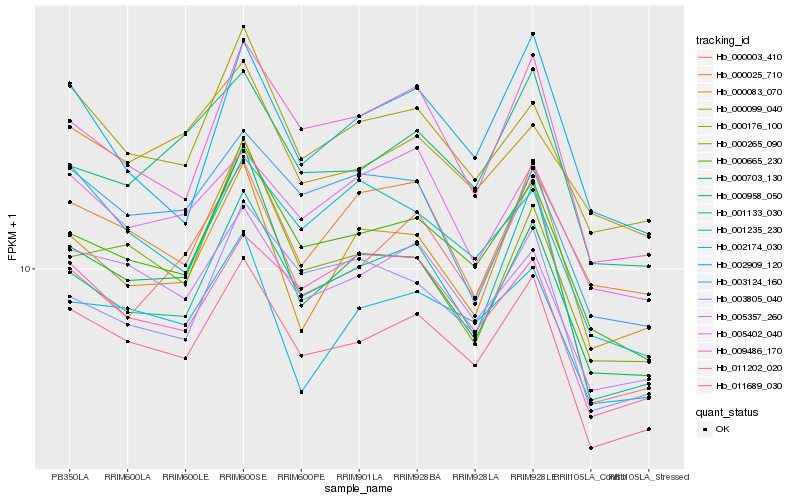
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011689_030 |
0.0 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 2 |
Hb_000665_230 |
0.0434022114 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 3 |
Hb_005357_260 |
0.0456960885 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 4 |
Hb_000176_100 |
0.056682363 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 5 |
Hb_001133_030 |
0.0592509887 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 6 |
Hb_009486_170 |
0.0607627835 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 7 |
Hb_000958_050 |
0.0630073839 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 8 |
Hb_002174_030 |
0.0631401576 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 9 |
Hb_011202_020 |
0.0674318807 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000703_130 |
0.0689689692 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 11 |
Hb_003805_040 |
0.0712310688 |
- |
- |
Sacsin [Glycine soja] |
| 12 |
Hb_000025_710 |
0.0739997727 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 13 |
Hb_005402_040 |
0.0741250239 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 14 |
Hb_001235_230 |
0.0743063454 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 15 |
Hb_000099_040 |
0.0748436233 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_000265_090 |
0.0758078611 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 17 |
Hb_003124_160 |
0.0758187116 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_002909_120 |
0.0763026806 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 19 |
Hb_000003_410 |
0.0764939517 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 20 |
Hb_000083_070 |
0.0766493767 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |